Orchestral maneuvers at the damaged sites in nucleotide excision repair
- PMID: 25681868
- PMCID: PMC11113351
- DOI: 10.1007/s00018-015-1859-5
Orchestral maneuvers at the damaged sites in nucleotide excision repair
Abstract
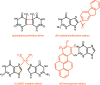
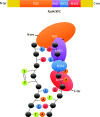
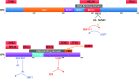
To safeguard the genome from the accumulation of deleterious effects arising from DNA lesions, cells developed several DNA repair mechanisms that remove specific types of damage from the genome. Among them, Nucleotide Excision Repair (NER) is unique in its ability to remove a very broad spectrum of lesions, the most important of which include UV-induced damage, bulky chemical adducts and some forms of oxidative damage. Two sub-pathways exist in NER; Transcription-Coupled Repair (TC-NER) removes lesion localized exclusively in transcribed genes while Global Genome Repair (GG-NER) removes lesions elsewhere. In TC- or GG-NER, more than 30 proteins detect, open, incise and resynthesize DNA. Intriguingly, half of them are involved in the detection of DNA damage, implying that this is a crucial repair step requiring a high level of regulation. We review here the complex damage recognition step of GG-NER with a focus on post-translational modifications that help the comings and goings of several protein complexes on the same short damaged DNA locus.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

