A novel statistical method for quantitative comparison of multiple ChIP-seq datasets
- PMID: 25682068
- PMCID: PMC4542775
- DOI: 10.1093/bioinformatics/btv094
A novel statistical method for quantitative comparison of multiple ChIP-seq datasets
Abstract
Motivation: ChIP-seq is a powerful technology to measure the protein binding or histone modification strength in the whole genome scale. Although there are a number of methods available for single ChIP-seq data analysis (e.g. 'peak detection'), rigorous statistical method for quantitative comparison of multiple ChIP-seq datasets with the considerations of data from control experiment, signal to noise ratios, biological variations and multiple-factor experimental designs is under-developed.
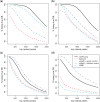
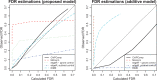
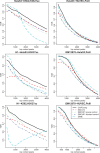
Results: In this work, we develop a statistical method to perform quantitative comparison of multiple ChIP-seq datasets and detect genomic regions showing differential protein binding or histone modification. We first detect peaks from all datasets and then union them to form a single set of candidate regions. The read counts from IP experiment at the candidate regions are assumed to follow Poisson distribution. The underlying Poisson rates are modeled as an experiment-specific function of artifacts and biological signals. We then obtain the estimated biological signals and compare them through the hypothesis testing procedure in a linear model framework. Simulations and real data analyses demonstrate that the proposed method provides more accurate and robust results compared with existing ones.
Availability and implementation: An R software package ChIPComp is freely available at http://web1.sph.emory.edu/users/hwu30/software/ChIPComp.html.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures


Similar articles
-
Using combined evidence from replicates to evaluate ChIP-seq peaks.Bioinformatics. 2015 Sep 1;31(17):2761-9. doi: 10.1093/bioinformatics/btv293. Epub 2015 May 7. Bioinformatics. 2015. PMID: 25957351
-
A signal-noise model for significance analysis of ChIP-seq with negative control.Bioinformatics. 2010 May 1;26(9):1199-204. doi: 10.1093/bioinformatics/btq128. Epub 2010 Apr 5. Bioinformatics. 2010. PMID: 20371496
-
BroadPeak: a novel algorithm for identifying broad peaks in diffuse ChIP-seq datasets.Bioinformatics. 2013 Feb 15;29(4):492-3. doi: 10.1093/bioinformatics/bts722. Epub 2013 Jan 7. Bioinformatics. 2013. PMID: 23300134
-
A short survey of computational analysis methods in analysing ChIP-seq data.Hum Genomics. 2011 Jan;5(2):117-23. doi: 10.1186/1479-7364-5-2-117. Hum Genomics. 2011. PMID: 21296745 Free PMC article. Review.
-
[Processing and analysis of ChIP-seq data].Yi Chuan. 2012 Jun;34(6):773-83. doi: 10.3724/sp.j.1005.2012.00773. Yi Chuan. 2012. PMID: 22698750 Review. Chinese.
Cited by
-
scaDA: A Novel Statistical Method for Differential Analysis of Single-Cell Chromatin Accessibility Sequencing Data.bioRxiv [Preprint]. 2024 Jan 24:2024.01.21.576570. doi: 10.1101/2024.01.21.576570. bioRxiv. 2024. Update in: PLoS Comput Biol. 2024 Aug 2;20(8):e1011854. doi: 10.1371/journal.pcbi.1011854. PMID: 38328112 Free PMC article. Updated. Preprint.
-
Comprehensive assessment of differential ChIP-seq tools guides optimal algorithm selection.Genome Biol. 2022 May 24;23(1):119. doi: 10.1186/s13059-022-02686-y. Genome Biol. 2022. PMID: 35606795 Free PMC article.
-
Recent advances in ChIP-seq analysis: from quality management to whole-genome annotation.Brief Bioinform. 2017 Mar 1;18(2):279-290. doi: 10.1093/bib/bbw023. Brief Bioinform. 2017. PMID: 26979602 Free PMC article. Review.
-
chromswitch: a flexible method to detect chromatin state switches.Bioinformatics. 2018 Jul 1;34(13):2286-2288. doi: 10.1093/bioinformatics/bty075. Bioinformatics. 2018. PMID: 29438498 Free PMC article.
-
scaDA: A novel statistical method for differential analysis of single-cell chromatin accessibility sequencing data.PLoS Comput Biol. 2024 Aug 2;20(8):e1011854. doi: 10.1371/journal.pcbi.1011854. eCollection 2024 Aug. PLoS Comput Biol. 2024. PMID: 39093856 Free PMC article.
References
-
- Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodological), 57, 289–300.
-
- Chen X., et al. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell , 133, 1106–1117. - PubMed
-
- Efron B. (2004) Large-scale simultaneous hypothesis testing. J. Am. Stat. Assoc. , 99, 96–104.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

