The Scalable Brain Atlas: Instant Web-Based Access to Public Brain Atlases and Related Content
- PMID: 25682754
- PMCID: PMC4469098
- DOI: 10.1007/s12021-014-9258-x
The Scalable Brain Atlas: Instant Web-Based Access to Public Brain Atlases and Related Content
Abstract
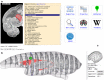
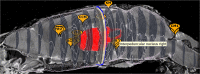
The Scalable Brain Atlas (SBA) is a collection of web services that provide unified access to a large collection of brain atlas templates for different species. Its main component is an atlas viewer that displays brain atlas data as a stack of slices in which stereotaxic coordinates and brain regions can be selected. These are subsequently used to launch web queries to resources that require coordinates or region names as input. It supports plugins which run inside the viewer and respond when a new slice, coordinate or region is selected. It contains 20 atlas templates in six species, and plugins to compute coordinate transformations, display anatomical connectivity and fiducial points, and retrieve properties, descriptions, definitions and 3d reconstructions of brain regions. The ambition of SBA is to provide a unified representation of all publicly available brain atlases directly in the web browser, while remaining a responsive and light weight resource that specializes in atlas comparisons, searches, coordinate transformations and interactive displays.
Conflict of interest statement
The authors have no conflict of interest
Figures





Similar articles
-
Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system.Nucleic Acids Res. 2013 Jan;41(Database issue):D996-D1008. doi: 10.1093/nar/gks1042. Epub 2012 Nov 28. Nucleic Acids Res. 2013. PMID: 23193282 Free PMC article.
-
Brain templates and atlases.Neuroimage. 2012 Aug 15;62(2):911-22. doi: 10.1016/j.neuroimage.2012.01.024. Epub 2012 Jan 10. Neuroimage. 2012. PMID: 22248580 Review.
-
AtlasGuide: software for stereotaxic guidance using 3D CT/MRI hybrid atlases of developing mouse brains.J Neurosci Methods. 2013 Oct 30;220(1):75-84. doi: 10.1016/j.jneumeth.2013.08.017. Epub 2013 Aug 27. J Neurosci Methods. 2013. PMID: 23994359 Free PMC article.
-
A hybrid approach to shape-based interpolation of stereotactic atlases of the human brain.Neuroinformatics. 2006;4(2):177-98. doi: 10.1385/NI:4:2:177. Neuroinformatics. 2006. PMID: 16845168
-
Discrepancies in stereotaxic coordinate publications and improving precision using an animal-specific atlas.J Neurosci Methods. 2017 Jun 1;284:15-20. doi: 10.1016/j.jneumeth.2017.03.019. Epub 2017 Apr 7. J Neurosci Methods. 2017. PMID: 28392415 Review.
Cited by
-
Ultrasound localization microscopy and functional ultrasound imaging reveal atypical features of the trigeminal ganglion vasculature.Commun Biol. 2022 Apr 7;5(1):330. doi: 10.1038/s42003-022-03273-4. Commun Biol. 2022. PMID: 35393515 Free PMC article.
-
In vivo dynamics and anti-tumor effects of EpCAM-directed CAR T-cells against brain metastases from lung cancer.Oncoimmunology. 2023 Jan 13;12(1):2163781. doi: 10.1080/2162402X.2022.2163781. eCollection 2023. Oncoimmunology. 2023. PMID: 36687005 Free PMC article.
-
Pten haploinsufficiency causes desynchronized growth of brain areas involved in sensory processing.iScience. 2022 Jan 19;25(2):103796. doi: 10.1016/j.isci.2022.103796. eCollection 2022 Feb 18. iScience. 2022. PMID: 35198865 Free PMC article.
-
Flexible, Penetrating Brain Probes Enabled by Advances in Polymer Microfabrication.Micromachines (Basel). 2016 Oct 4;7(10):180. doi: 10.3390/mi7100180. Micromachines (Basel). 2016. PMID: 30404353 Free PMC article. Review.
-
Replenished microglia partially rescue schizophrenia-related stress response.Front Cell Neurosci. 2023 Sep 12;17:1254923. doi: 10.3389/fncel.2023.1254923. eCollection 2023. Front Cell Neurosci. 2023. PMID: 37771931 Free PMC article.
References
-
- Bray, T., Paoli, J., Sperberg-McQueen, C. M., Maler, E., & Yergeau, F. (Eds.) (2008) “Extensible Markup Language (XML) 1.0”, W3C recommendation http://www.w3.org/TR/2008/REC-xml-20081126/.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

