A Golgi-based KDELR-dependent signalling pathway controls extracellular matrix degradation
- PMID: 25682866
- PMCID: PMC4413660
- DOI: 10.18632/oncotarget.3270
A Golgi-based KDELR-dependent signalling pathway controls extracellular matrix degradation
Abstract
We recently identified an endomembrane-based signalling cascade that is activated by the KDEL receptor (KDELR) on the Golgi complex. At the Golgi, the KDELR acts as a traffic sensor (presumably via binding to chaperones that leave the ER) and triggers signalling pathways that balance membrane fluxes between ER and Golgi. One such pathway relies on Gq and Src. Here, we examine if KDELR might control other cellular modules through this pathway. Given the central role of Src in extracellular matrix (ECM) degradation, we investigated the impact of the KDELR-Src pathway on the ability of cancer cells to degrade the ECM. We find that activation of the KDELR controls ECM degradation by increasing the number of the degradative structures known as invadopodia. The KDELR induces Src activation at the invadopodia and leads to phosphorylation of the Src substrates cortactin and ASAP1, which are required for basal and KDELR-stimulated ECM degradation. This study furthers our understanding of the regulatory circuitry underlying invadopodia-dependent ECM degradation, a key phase in metastases formation and invasive growth.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
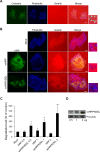



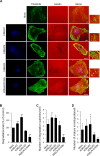
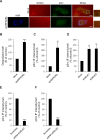

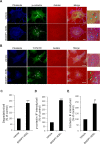

References
-
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–52. - PubMed
-
- Csete ME, Doyle JC. Reverse engineering of biological complexity. Science. 2002;295:1664–1669. - PubMed
-
- Sontag ED. Molecular systems biology and control. Eur J Control. 2005;11:396–435.
-
- Cancino J, Capalbo A, Di Campli A, Giannotta M, Rizzo R, Jung JE, Di Martino R, Persico M, Heinklein P, Sallese M, Luini A. Control Systems of Membrane Transport at the Interface between the Endoplasmic Reticulum and the Golgi. Dev Cell. 2014;30:280–294. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

