Initiation and regulation of paramyxovirus transcription and replication
- PMID: 25683441
- PMCID: PMC4424093
- DOI: 10.1016/j.virol.2015.01.014
Initiation and regulation of paramyxovirus transcription and replication
Abstract
The paramyxovirus family has a genome consisting of a single strand of negative sense RNA. This genome acts as a template for two distinct processes: transcription to generate subgenomic, capped and polyadenylated mRNAs, and genome replication. These viruses only encode one polymerase. Thus, an intriguing question is, how does the viral polymerase initiate and become committed to either transcription or replication? By answering this we can begin to understand how these two processes are regulated. In this review article, we present recent findings from studies on the paramyxovirus, respiratory syncytial virus, which show how its polymerase is able to initiate transcription and replication from a single promoter. We discuss how these findings apply to other paramyxoviruses. Then, we examine how trans-acting proteins and promoter secondary structure might serve to regulate transcription and replication during different phases of the paramyxovirus replication cycle.
Keywords: Gene expression; Mononegavirales; Non-segmented negative strand RNA virus; Paramyxoviridae; Paramyxovirus; Polymerase; Promoter; Replication; Transcription.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures


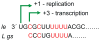
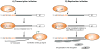

References
-
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus taxonomy: classification and nomenclature of viruses: Ninthe Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; 2012.
-
- Lamb RA, Parks GD. Paramyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams and Wilkins; pp. 1449–1496.
-
- Whelan SP, Barr JN, Wertz GW. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol. 2004;283:61–119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

