UCR1C is a novel activator of phosphodiesterase 4 (PDE4) long isoforms and attenuates cardiomyocyte hypertrophy
- PMID: 25683917
- PMCID: PMC4361369
- DOI: 10.1016/j.cellsig.2015.02.003
UCR1C is a novel activator of phosphodiesterase 4 (PDE4) long isoforms and attenuates cardiomyocyte hypertrophy
Abstract
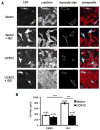
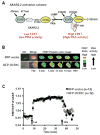
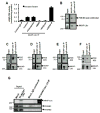
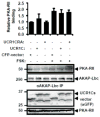
Hypertrophy increases the risk of heart failure and arrhythmia. Prevention or reversal of the maladaptive hypertrophic phenotype has thus been proposed to treat heart failure. Chronic β-adrenergic receptor (β-AR) stimulation induces cardiomyocyte hypertrophy by elevating 3',5'-cyclic adenosine monophosphate (cAMP) levels and activating downstream effectors such protein kinase A (PKA). Conversely, hydrolysis of cAMP by phosphodiesterases (PDEs) spatiotemporally restricts cAMP signaling. Here, we demonstrate that PDE4, but not PDE3, is critical in regulating cardiomyocyte hypertrophy, and may represent a potential target for preventing maladaptive hypertrophy. We identify a sequence within the upstream conserved region 1 of PDE4D, termed UCR1C, as a novel activator of PDE4 long isoforms. UCR1C activates PDE4 in complex with A-kinase anchoring protein (AKAP)-Lbc resulting in decreased PKA signaling facilitated by AKAP-Lbc. Expression of UCR1C in cardiomyocytes inhibits hypertrophy in response to chronic β-AR stimulation. This effect is partially due to inhibition of nuclear PKA activity, which decreases phosphorylation of the transcription factor cAMP response element-binding protein (CREB). In conclusion, PDE4 activation by UCR1C attenuates cardiomyocyte hypertrophy by specifically inhibiting nuclear PKA activity.
Keywords: 3′,5′-cyclic monophosphate (cAMP); A-kinase anchoring protein (AKAP); Compartmentalized signaling; Phosphodiesterase 4 (PDE4) activation; cardiomyocyte hypertrophy; protein kinase A (PKA).
Published by Elsevier Inc.
Conflict of interest statement
None declared
Figures








References
-
- Hill JA, Olson EN. The New England journal of medicine. 2008;358:1370–1380. - PubMed
-
- Selvetella G, Hirsch E, Notte A, Tarone G, Lembo G. Cardiovascular research. 2004;63:373–380. - PubMed
-
- Frey N, Katus HA, Olson EN, Hill JA. Circulation. 2004;109:1580–1589. - PubMed
-
- Frey N, Olson EN. Annual review of physiology. 2003;65:45–79. - PubMed
-
- Heineke J, Molkentin JD. Nature reviews Molecular cell biology. 2006;7:589–600. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

