Stable oncogenic silencing in vivo by programmable and targeted de novo DNA methylation in breast cancer
- PMID: 25684141
- PMCID: PMC4633433
- DOI: 10.1038/onc.2014.470
Stable oncogenic silencing in vivo by programmable and targeted de novo DNA methylation in breast cancer
Abstract
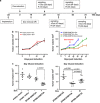
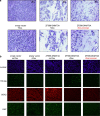
With the recent comprehensive mapping of cancer genomes, there is now a need for functional approaches to edit the aberrant epigenetic state of key cancer drivers to reprogram the epi-pathology of the disease. In this study we utilized a programmable DNA-binding methyltransferase to induce targeted incorporation of DNA methylation (DNAme) in the SOX2 oncogene in breast cancer through a six zinc finger (ZF) protein linked to DNA methyltransferase 3A (ZF-DNMT3A). We demonstrated long-lasting oncogenic repression, which was maintained even after suppression of ZF-DNMT3A expression in tumor cells. The de novo DNAme was faithfully propagated and maintained through cell generations even after the suppression of the expression of the chimeric methyltransferase in the tumor cells. Xenograft studies in NUDE mice demonstrated stable SOX2 repression and long-term breast tumor growth inhibition, which lasted for >100 days post implantation of the tumor cells in mice. This was accompanied with a faithful maintenance of DNAme in the breast cancer implants. In contrast, downregulation of SOX2 by ZF domains engineered with the Krueppel-associated box repressor domain resulted in a transient and reversible suppression of oncogenic gene expression. Our results indicated that targeted de novo DNAme of the SOX2 oncogenic promoter was sufficient to induce long-lasting epigenetic silencing, which was not only maintained during cell division but also significantly delayed the tumorigenic phenotype of cancer cells in vivo, even in the absence of treatment. Here, we outline a genome-based targeting approach to long-lasting tumor growth inhibition with potential applicability to many other oncogenic drivers that are currently refractory to drug design.
Figures



References
-
- 1Kouzarides T. Chromatin modifications and their function. Cell 2007; 128: 693–705. - PubMed
-
- 2Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet 2011; 12: 123–135. - PubMed
-
- 3Lange UC, Schneider R. What an epigenome remembers. Bioessays 2010; 32: 659–668. - PubMed
-
- 4Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev 2012; 22: 50–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

