DNA Methylation and Histone Acetylation Patterns in Cultured Bovine Adipose Tissue-Derived Stem Cells (BADSCs)
- PMID: 25685737
- PMCID: PMC4297485
- DOI: 10.22074/cellj.2015.492
DNA Methylation and Histone Acetylation Patterns in Cultured Bovine Adipose Tissue-Derived Stem Cells (BADSCs)
Abstract
Objective: Many studies have focused on the epigenetic characteristics of donor cells to improve somatic cell nuclear transfer (SCNT). We hypothesized that the epigenetic status and chromatin structure of undifferentiated bovine adipose tissue-derived stem cells (BADSCs) would not remain constant during different passages. The objective of this study was to determine the mRNA expression patterns of DNA methyltransferases (DNMT1, DNMT3a, DNMT3b) and histone deacetyltransferses (HDAC1, HDAC2, HDAC3) in BADSCs. In addition, we compared the measured levels of octamer binding protein-4 expression (OCT4) and acetylation of H3K9 (H3K9ac) in BADSCs cultures and different passages in vitro.
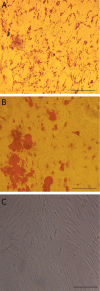
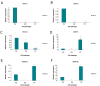
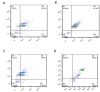
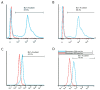
Materials and methods: In this experimental study, subcutaneous fat was obtained from adult cows immediately post-mortem. Relative level of DNMTs and HDACs was examined using quantitative real time polymerase chain reaction (q-PCR), and the level of OCT4 and H3K9ac was analyzed by flow cytometry at passages 3 (P3), 5 (P5) and 7 (P7).
Results: The OCT4 protein level was similar at P3 and P5 but a significant decrease in its level was seen at P7. The highest and lowest levels of H3K9ac were observed at P5 and P7, respectively. At P5, the expression of HDACs and DNMTs was significantly decreased. In contrast, a remarkable increase in the expression of DNMTs was observed at P7.
Conclusion: Our data demonstrated that the epigenetic status of BADSCs was variable during culture. The P5 cells showed the highest level of stemness and multipotency and the lowest level of chromatin compaction. Therefore, we suggest that P5 cells may be more efficient for SCNT compared with other passages.
Keywords: DNA Methyltransferases; Epigenetics; Histone Deacetyltransferses; Somatic Cell Nuclear Transfer.
Figures
References
-
- Hill JR. Abnormal in utero development of cloned animals: implications for human cloning. Differentiation. 2002;69(4-5):174–178. - PubMed
-
- Cezar GG. Epigenetic reprogramming of cloned animals. Cloning Stem Cells. 2003;5(3):165–180. - PubMed
-
- Giraldo AM, Lynn JW, Purpera MN, Godke RA, Bondioli KR. DNA methylation and histone acetylation patterns in cultured bovine fibroblasts for nuclear transfer. Mol Reprod Dev. 2007;74(12):1514–1524. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
