Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow
- PMID: 25687283
- PMCID: PMC4354373
- DOI: 10.1084/jem.20141442
Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow
Abstract
In mice, two restricted dendritic cell (DC) progenitors, macrophage/dendritic progenitors (MDPs) and common dendritic progenitors (CDPs), demonstrate increasing commitment to the DC lineage, as they sequentially lose granulocyte and monocyte potential, respectively. Identifying these progenitors has enabled us to understand the role of DCs and monocytes in immunity and tolerance in mice. In humans, however, restricted monocyte and DC progenitors remain unknown. Progress in studying human DC development has been hampered by lack of an in vitro culture system that recapitulates in vivo DC hematopoiesis. Here we report a culture system that supports development of CD34(+) hematopoietic stem cell progenitors into the three major human DC subsets, monocytes, granulocytes, and NK and B cells. Using this culture system, we defined the pathway for human DC development and revealed the sequential origin of human DCs from increasingly restricted progenitors: a human granulocyte-monocyte-DC progenitor (hGMDP) that develops into a human monocyte-dendritic progenitor (hMDP), which in turn develops into monocytes, and a human CDP (hCDP) that is restricted to produce the three major DC subsets. The phenotype of the DC progenitors partially overlaps with granulocyte-macrophage progenitors (GMPs). These progenitors reside in human cord blood and bone marrow but not in the blood or lymphoid tissues.
© 2015 Lee et al.
Figures

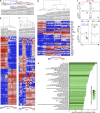



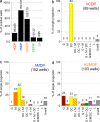


Comment in
-
The real thing: how to make human DC subsets.J Exp Med. 2015 Mar 9;212(3):285. doi: 10.1084/jem.2123insight2. J Exp Med. 2015. PMID: 25754119 Free PMC article. No abstract available.
References
-
- Boissel N., Rousselot P., Raffoux E., Cayuela J.M., Maarek O., Charron D., Degos L., Dombret H., Toubert A., and Rea D.. 2004. Defective blood dendritic cells in chronic myeloid leukemia correlate with high plasmatic VEGF and are not normalized by imatinib mesylate. Leukemia. 18:1656–1661 10.1038/sj.leu.2403474 - DOI - PubMed
-
- Chang C.C., Wright A., and Punnonen J.. 2000. Monocyte-derived CD1a+ and CD1a− dendritic cell subsets differ in their cytokine production profiles, susceptibilities to transfection, and capacities to direct Th cell differentiation. J. Immunol. 165:3584–3591 10.4049/jimmunol.165.7.3584 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

