High-resolution crystal structure of Z-DNA in complex with Cr(3+) cations
- PMID: 25687556
- PMCID: PMC4381091
- DOI: 10.1007/s00775-015-1247-5
High-resolution crystal structure of Z-DNA in complex with Cr(3+) cations
Abstract
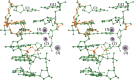
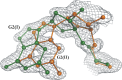
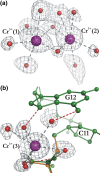
This work is part of our project aimed at characterizing metal-binding properties of left-handed Z-DNA helices. The three Cr(3+) cations found in the asymmetric unit of the d(CGCGCG)2-Cr(3+) crystal structure do not form direct coordination bonds with atoms of the Z-DNA molecule. Instead, the hydrated Cr(3+) ions are engaged in outer-sphere interactions with phosphate groups and O6 and N7 guanine atoms of the DNA. The Cr(3+)(1) and Cr(3+)(2) ions have disordered coordination spheres occupied by six water molecules each. These partial-occupancy chromium cations are 2.354(15) Å apart and are bridged by three water molecules from their hydration spheres. The Cr(3+)(3) cation has distorted square pyramidal geometry. In addition to the high degree of disorder of the DNA backbone, alternate conformations are also observed for the deoxyribose and base moieties of the G2 nucleotide. Our work illuminates the question of conformational flexibility of Z-DNA and its interaction mode with transition-metal cations.
Figures
Similar articles
-
Crystal structure of Z-DNA in complex with the polyamine putrescine and potassium cations at ultra-high resolution.Acta Crystallogr B Struct Sci Cryst Eng Mater. 2021 Jun 1;77(Pt 3):331-338. doi: 10.1107/S2052520621002663. Epub 2021 May 6. Acta Crystallogr B Struct Sci Cryst Eng Mater. 2021. PMID: 34096514 Free PMC article.
-
Ultrahigh-resolution crystal structures of Z-DNA in complex with Mn(2+) and Zn(2+) ions.Acta Crystallogr D Biol Crystallogr. 2013 Jun;69(Pt 6):1180-90. doi: 10.1107/S0907444913007798. Epub 2013 May 16. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 23695262
-
Ultrahigh-resolution centrosymmetric crystal structure of Z-DNA reveals the massive presence of alternate conformations.Acta Crystallogr D Struct Biol. 2016 Nov 1;72(Pt 11):1203-1211. doi: 10.1107/S205979831601679X. Epub 2016 Oct 28. Acta Crystallogr D Struct Biol. 2016. PMID: 27841753
-
Crystal structures of A-DNA duplexes.Biopolymers. 1997;44(1):45-63. doi: 10.1002/(SICI)1097-0282(1997)44:1<45::AID-BIP4>3.0.CO;2-#. Biopolymers. 1997. PMID: 9097733 Review.
-
Cations as hydrogen bond donors: a view of electrostatic interactions in DNA.Annu Rev Biophys Biomol Struct. 2003;32:27-45. doi: 10.1146/annurev.biophys.32.110601.141726. Epub 2003 Feb 14. Annu Rev Biophys Biomol Struct. 2003. PMID: 12598364 Review.
Cited by
-
Conformation-dependent restraints for polynucleotides: the sugar moiety.Nucleic Acids Res. 2020 Jan 24;48(2):962-973. doi: 10.1093/nar/gkz1122. Nucleic Acids Res. 2020. PMID: 31799624 Free PMC article.
-
Polarizable Force Field for DNA Based on the Classical Drude Oscillator: II. Microsecond Molecular Dynamics Simulations of Duplex DNA.J Chem Theory Comput. 2017 May 9;13(5):2072-2085. doi: 10.1021/acs.jctc.7b00068. Epub 2017 Apr 19. J Chem Theory Comput. 2017. PMID: 28398748 Free PMC article.
-
Crystal structure of Z-DNA in complex with the polyamine putrescine and potassium cations at ultra-high resolution.Acta Crystallogr B Struct Sci Cryst Eng Mater. 2021 Jun 1;77(Pt 3):331-338. doi: 10.1107/S2052520621002663. Epub 2021 May 6. Acta Crystallogr B Struct Sci Cryst Eng Mater. 2021. PMID: 34096514 Free PMC article.
-
Nonalternating purine pyrimidine sequences can form stable left-handed DNA duplex by strong topological constraint.Nucleic Acids Res. 2022 Jan 25;50(2):684-696. doi: 10.1093/nar/gkab1283. Nucleic Acids Res. 2022. PMID: 34967416 Free PMC article.
-
Do Ternary DNA-Cr(III)-Small Molecule Adducts Form?Biol Trace Elem Res. 2025 May 1:10.1007/s12011-025-04641-x. doi: 10.1007/s12011-025-04641-x. Online ahead of print. Biol Trace Elem Res. 2025. PMID: 40312602 Free PMC article.
References
-
- Cieslak-Golonka M. Polyhedron. 1995;15:3667–3689. doi: 10.1016/0277-5387(96)00141-6. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

