Chromatin compaction under mixed salt conditions: opposite effects of sodium and potassium ions on nucleosome array folding
- PMID: 25688036
- PMCID: PMC4330525
- DOI: 10.1038/srep08512
Chromatin compaction under mixed salt conditions: opposite effects of sodium and potassium ions on nucleosome array folding
Abstract
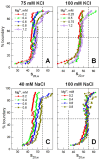
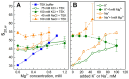
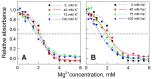
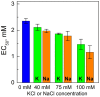
It is well known that chromatin structure is highly sensitive to the ionic environment. However, the combined effects of a physiologically relevant mixed ionic environment of K(+), Mg(2+) and Na(+), which are the main cations of the cell cytoplasm, has not been systematically investigated. We studied folding and self-association (aggregation) of recombinant 12-mer nucleosome arrays with 177 bp DNA repeat length in solutions of mixtures of K(+) and Mg(2+) or Na(+) and Mg(2+). In the presence of Mg(2+), the addition of sodium ions promotes folding of array into 30-nm fibres, whereas in mixtures of K(+) and Mg(2+), potassium ions abrogate folding. We found that self-association of nucleosome arrays in mixed salt solutions is synergistically promoted by Mg(2+) and monovalent ions, with sodium being slightly more efficient than potassium in amplifying the self-association. The results highlight the importance of a mixed ionic environment for the compaction of chromatin under physiological conditions and demonstrate the complicated nature of the various factors that determine and regulate chromatin compaction in vivo.
Figures
References
-
- Wolffe A. P. Chromatin: Structure and function, 447 (Academic Press, San DiegoCA, 1998).
-
- Grigoryev S. A. & Woodcock C. L. Chromatin organization - The 30 nm fiber. Exp Cell Res. 318, 1448–1455 (2012). - PubMed
-
- Woodcock C. L., Skoultchi A. I. & Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 14, 17–25 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

