Sulfide oxidation by a noncanonical pathway in red blood cells generates thiosulfate and polysulfides
- PMID: 25688092
- PMCID: PMC4375485
- DOI: 10.1074/jbc.M115.639831
Sulfide oxidation by a noncanonical pathway in red blood cells generates thiosulfate and polysulfides
Abstract
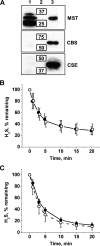
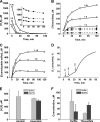
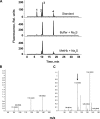
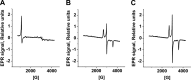
A cardioprotectant at low concentrations, H2S is a toxin at high concentrations and inhibits cytochrome c oxidase. A conundrum in H2S homeostasis is its fate in red blood cells (RBCs), which produce H2S but lack the canonical mitochondrial sulfide oxidation pathway for its clearance. The sheer abundance of RBCs in circulation enhances the metabolic significance of their clearance strategy for H2S, necessary to avoid systemic toxicity. In this study, we demonstrate that H2S generation by RBCs is catalyzed by mercaptopyruvate sulfurtransferase. Furthermore, we have discovered the locus of sulfide oxidation in RBCs and describe a new role for an old protein, hemoglobin, which in the ferric or methemoglobin state binds H2S and oxidizes it to a mixture of thiosulfate and hydropolysulfides. Our study reveals a previously undescribed route for the biogenesis of hydropolysulfides, which are increasingly considered important for H2S-based signaling, but their origin in mammalian cells is unknown. An NADPH/flavoprotein oxidoreductase system restores polysulfide-carrying hemoglobin derivatives to ferrous hemoglobin, thus completing the methemoglobin-dependent sulfide oxidation cycle. Methemoglobin-dependent sulfide oxidation in mammals is complex and has similarities to chemistry reported for the dissolution of iron oxides in sulfidic waters and during bioleaching of metal sulfides. The catalytic oxidation of H2S by hemoglobin explains how RBCs maintain low steady-state H2S levels in circulation, and suggests that additional hemeproteins might be involved in sulfide homeostasis in other tissues.
Keywords: Heme; Hemoglobin; Hydrogen Sulfide; Oxidation-Reduction (Redox); Sulfur.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Theissen U., Hoffmeister M., Grieshaber M., Martin W. (2003) Single eubacterial origin of eukaryotic sulfide:quinone oxidoreductase, a mitochondrial enzyme conserved from the early evolution of eukaryotes during anoxic and sulfidic times. Mol. Biol. Evol. 20, 1564–1574 - PubMed
-
- Riahi S., Rowley C. N. (2014) Why can hydrogen sulfide permeate cell membranes? J Am. Chem. Soc. 136, 15111–15113 - PubMed
-
- Kimura H. (2010) Hydrogen sulfide: from brain to gut. Antioxid. Redox Signal. 12, 1111–1123 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

