Global DNA cytosine methylation as an evolving trait: phylogenetic signal and correlated evolution with genome size in angiosperms
- PMID: 25688257
- PMCID: PMC4310347
- DOI: 10.3389/fgene.2015.00004
Global DNA cytosine methylation as an evolving trait: phylogenetic signal and correlated evolution with genome size in angiosperms
Abstract
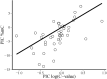
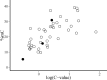
DNA cytosine methylation is a widespread epigenetic mechanism in eukaryotes, and plant genomes commonly are densely methylated. Genomic methylation can be associated with functional consequences such as mutational events, genomic instability or altered gene expression, but little is known on interspecific variation in global cytosine methylation in plants. In this paper, we compare global cytosine methylation estimates obtained by HPLC and use a phylogenetically-informed analytical approach to test for significance of evolutionary signatures of this trait across 54 angiosperm species in 25 families. We evaluate whether interspecific variation in global cytosine methylation is statistically related to phylogenetic distance and also whether it is evolutionarily correlated with genome size (C-value). Global cytosine methylation varied widely between species, ranging between 5.3% (Arabidopsis) and 39.2% (Narcissus). Differences between species were related to their evolutionary trajectories, as denoted by the strong phylogenetic signal underlying interspecific variation. Global cytosine methylation and genome size were evolutionarily correlated, as revealed by the significant relationship between the corresponding phylogenetically independent contrasts. On average, a ten-fold increase in genome size entailed an increase of about 10% in global cytosine methylation. Results show that global cytosine methylation is an evolving trait in angiosperms whose evolutionary trajectory is significantly linked to changes in genome size, and suggest that the evolutionary implications of epigenetic mechanisms are likely to vary between plant lineages.
Keywords: C-value; DNA cytosine methylation; HPLC; angiosperms; correlated evolution; epigenetics; genome size; phylogenetic signal.
Figures
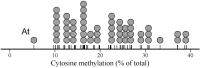

References
-
- Banks J., Federoff N. (1989). Patterns of developmental and heritable change in methylation of the suppressor-mutator transposable element. Dev. Genet. 10, 425–437 10.1002/dvg.1020100604 - DOI
-
- Bennett M., Leitch I. (2005). Genome size evolution in plants, in The Evolution of the Genome, ed Gregory T. R. (Amsterdam: Elsevier; ), 89–162.
-
- Bennett M., Leitch I. (2012). Angiosperm DNA C-values Database (Release 8.0, December 2012). Available online at: http://www.kew.org/cvalues/
LinkOut - more resources
Full Text Sources
Other Literature Sources

