Long-Range PCR Amplification of DNA by DNA Polymerase III Holoenzyme from Thermus thermophilus
- PMID: 25688300
- PMCID: PMC4320859
- DOI: 10.1155/2015/837842
Long-Range PCR Amplification of DNA by DNA Polymerase III Holoenzyme from Thermus thermophilus
Abstract
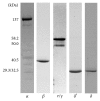
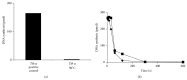
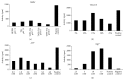
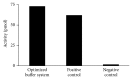
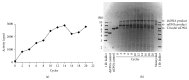
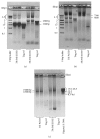
DNA replication in bacteria is accomplished by a multicomponent replicase, the DNA polymerase III holoenzyme (pol III HE). The three essential components of the pol III HE are the α polymerase, the β sliding clamp processivity factor, and the DnaX clamp-loader complex. We report here the assembly of the functional holoenzyme from Thermus thermophilus (Tth), an extreme thermophile. The minimal holoenzyme capable of DNA synthesis consists of α, β and DnaX (τ and γ), δ and δ' components of the clamp-loader complex. The proteins were each cloned and expressed in a native form. Each component of the system was purified extensively. The minimum holoenzyme from these five purified subunits reassembled is sufficient for rapid and processive DNA synthesis. In an isolated form the α polymerase was found to be unstable at temperatures above 65°C. We were able to increase the thermostability of the pol III HE to 98°C by addition and optimization of various buffers and cosolvents. In the optimized buffer system we show that a replicative polymerase apparatus, Tth pol III HE, is capable of rapid amplification of regions of DNA up to 15,000 base pairs in PCR reactions.
Figures


Similar articles
-
DNA polymerase III holoenzyme from Thermus thermophilus identification, expression, purification of components, and use to reconstitute a processive replicase.J Biol Chem. 2002 Apr 19;277(16):13401-8. doi: 10.1074/jbc.M110833200. Epub 2002 Jan 31. J Biol Chem. 2002. PMID: 11823461
-
Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. IV. ATP-binding site mutants identify the clamp loader.J Biol Chem. 1995 Jun 2;270(22):13378-83. doi: 10.1074/jbc.270.22.13378. J Biol Chem. 1995. PMID: 7768939
-
Total reconstitution of DNA polymerase III holoenzyme reveals dual accessory protein clamps.J Biol Chem. 1990 Jan 15;265(2):1179-87. J Biol Chem. 1990. PMID: 2404006
-
The E. coli DNA Replication Fork.Enzymes. 2016;39:31-88. doi: 10.1016/bs.enz.2016.04.001. Epub 2016 May 13. Enzymes. 2016. PMID: 27241927 Review.
-
DNA polymerase III holoenzyme of Escherichia coli: components and function of a true replicative complex.Mol Cell Biochem. 1985 Feb;66(1):71-85. doi: 10.1007/BF00231826. Mol Cell Biochem. 1985. PMID: 3885002 Review.
References
-
- Kornberg A., Baker T. A. DNA Replication. New York, NY, USA: W. H. Freeman; 1992.
-
- McHenry C. S. DNA polymerase III holoenzyme: components, structure, and mechanism of a true replicative complex. The Journal of Biological Chemistry. 1991;266(29):19127–19130. - PubMed
-
- LaDuca R. J., Crute J. J., McHenry C. S., Bambara R. A. The β subunit of the Escherichia coli DNA polymerase III holoenzyme interacts functionally with the catalytic core in the absence of other subunits. The Journal of Biological Chemistry. 1986;261(16):7550–7557. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
