Luminal progenitors restrict their lineage potential during mammary gland development
- PMID: 25688859
- PMCID: PMC4331521
- DOI: 10.1371/journal.pbio.1002069
Luminal progenitors restrict their lineage potential during mammary gland development
Abstract
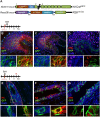
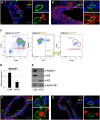
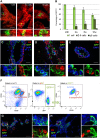
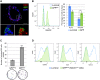
The hierarchical relationships between stem cells and progenitors that guide mammary gland morphogenesis are still poorly defined. While multipotent basal stem cells have been found within the myoepithelial compartment, the in vivo lineage potential of luminal progenitors is unclear. Here we used the expression of the Notch1 receptor, previously implicated in mammary gland development and tumorigenesis, to elucidate the hierarchical organization of mammary stem/progenitor cells by lineage tracing. We found that Notch1 expression identifies multipotent stem cells in the embryonic mammary bud, which progressively restrict their lineage potential during mammary ductal morphogenesis to exclusively generate an ERαneg luminal lineage postnatally. Importantly, our results show that Notch1-labelled cells represent the alveolar progenitors that expand during pregnancy and survive multiple successive involutions. This study reveals that postnatal luminal epithelial cells derive from distinct self-sustained lineages that may represent the cells of origin of different breast cancer subtypes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, et al. (2006) Generation of a functional mammary gland from a single stem cell. Nature 439: 84–88. - PubMed
-
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, et al. (2006) Purification and unique properties of mammary epithelial stem cells. Nature 439: 993–997. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

