Identification of modulators of the nuclear receptor peroxisome proliferator-activated receptor α (PPARα) in a mouse liver gene expression compendium
- PMID: 25689681
- PMCID: PMC4331523
- DOI: 10.1371/journal.pone.0112655
Identification of modulators of the nuclear receptor peroxisome proliferator-activated receptor α (PPARα) in a mouse liver gene expression compendium
Erratum in
-
Correction: identification of modulators of the nuclear receptor peroxisome proliferator-activated receptor α (PPARα) in a mouse liver gene expression compendium.PLoS One. 2015 Apr 7;10(4):e0124224. doi: 10.1371/journal.pone.0124224. eCollection 2015. PLoS One. 2015. PMID: 25849353 Free PMC article. No abstract available.
Abstract
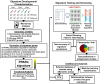
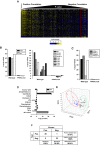
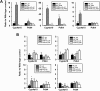
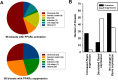
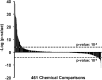
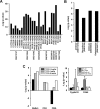
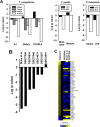
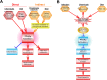
The nuclear receptor family member peroxisome proliferator-activated receptor α (PPARα) is activated by therapeutic hypolipidemic drugs and environmentally-relevant chemicals to regulate genes involved in lipid transport and catabolism. Chronic activation of PPARα in rodents increases liver cancer incidence, whereas suppression of PPARα activity leads to hepatocellular steatosis. Analytical approaches were developed to identify biosets (i.e., gene expression differences between two conditions) in a genomic database in which PPARα activity was altered. A gene expression signature of 131 PPARα-dependent genes was built using microarray profiles from the livers of wild-type and PPARα-null mice after exposure to three structurally diverse PPARα activators (WY-14,643, fenofibrate and perfluorohexane sulfonate). A fold-change rank-based test (Running Fisher's test (p-value ≤ 10(-4))) was used to evaluate the similarity between the PPARα signature and a test set of 48 and 31 biosets positive or negative, respectively for PPARα activation; the test resulted in a balanced accuracy of 98%. The signature was then used to identify factors that activate or suppress PPARα in an annotated mouse liver/primary hepatocyte gene expression compendium of ~1850 biosets. In addition to the expected activation of PPARα by fibrate drugs, di(2-ethylhexyl) phthalate, and perfluorinated compounds, PPARα was activated by benzofuran, galactosamine, and TCDD and suppressed by hepatotoxins acetaminophen, lipopolysaccharide, silicon dioxide nanoparticles, and trovafloxacin. Additional factors that activate (fasting, caloric restriction) or suppress (infections) PPARα were also identified. This study 1) developed methods useful for future screening of environmental chemicals, 2) identified chemicals that activate or suppress PPARα, and 3) identified factors including diets and infections that modulate PPARα activity and would be hypothesized to affect chemical-induced PPARα activity.
Conflict of interest statement
Figures


Similar articles
-
Identification of chemical modulators of the constitutive activated receptor (CAR) in a gene expression compendium.Nucl Recept Signal. 2015 Apr 27;13:e002. doi: 10.1621/nrs.13002. eCollection 2015. Nucl Recept Signal. 2015. PMID: 25949234 Free PMC article.
-
Screening a mouse liver gene expression compendium identifies modulators of the aryl hydrocarbon receptor (AhR).Toxicology. 2015 Oct 2;336:99-112. doi: 10.1016/j.tox.2015.07.005. Epub 2015 Jul 26. Toxicology. 2015. PMID: 26215100 Free PMC article.
-
Characterization of peroxisome proliferator-activated receptor alpha--independent effects of PPARalpha activators in the rodent liver: di-(2-ethylhexyl) phthalate also activates the constitutive-activated receptor.Toxicol Sci. 2010 Jan;113(1):45-59. doi: 10.1093/toxsci/kfp251. Epub 2009 Oct 22. Toxicol Sci. 2010. PMID: 19850644 Free PMC article.
-
Peroxisome proliferator-activated receptor-alpha and liver cancer: where do we stand?J Mol Med (Berl). 2005 Oct;83(10):774-85. doi: 10.1007/s00109-005-0678-9. Epub 2005 Jun 23. J Mol Med (Berl). 2005. PMID: 15976920 Review.
-
The PPARα-dependent rodent liver tumor response is not relevant to humans: addressing misconceptions.Arch Toxicol. 2018 Jan;92(1):83-119. doi: 10.1007/s00204-017-2094-7. Epub 2017 Dec 2. Arch Toxicol. 2018. PMID: 29197930 Free PMC article. Review.
Cited by
-
A gene expression biomarker for predictive toxicology to identify chemical modulators of NF-κB.PLoS One. 2022 Feb 2;17(2):e0261854. doi: 10.1371/journal.pone.0261854. eCollection 2022. PLoS One. 2022. PMID: 35108274 Free PMC article.
-
Toxicogenomics Approaches to Address Toxicity and Carcinogenicity in the Liver.Toxicol Pathol. 2023 Oct;51(7-8):470-481. doi: 10.1177/01926233241227942. Epub 2024 Jan 30. Toxicol Pathol. 2023. PMID: 38288963 Free PMC article. Review.
-
From the Cover: Genomic Effects of Androstenedione and Sex-Specific Liver Cancer Susceptibility in Mice.Toxicol Sci. 2017 Nov 1;160(1):15-29. doi: 10.1093/toxsci/kfx153. Toxicol Sci. 2017. PMID: 28973534 Free PMC article.
-
Identification of p53 Activators in a Human Microarray Compendium.Chem Res Toxicol. 2019 Sep 16;32(9):1748-1759. doi: 10.1021/acs.chemrestox.9b00052. Epub 2019 Sep 3. Chem Res Toxicol. 2019. PMID: 31397557 Free PMC article.
-
A set of six Gene expression biomarkers and their thresholds identify rat liver tumorigens in short-term assays.Toxicology. 2020 Oct;443:152547. doi: 10.1016/j.tox.2020.152547. Epub 2020 Aug 2. Toxicology. 2020. PMID: 32755643 Free PMC article.
References
-
- Peters JM, Cattley RC, Gonzalez FJ (1997) Role of PPAR alpha in the mechanism of action of the nongenotoxic carcinogen and peroxisome proliferator Wy-14,643. Carcinogenesis 18: 2029–2033. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

