The nucleoporin Mlp2 is involved in chromosomal distribution during mitosis in trypanosomatids
- PMID: 25690889
- PMCID: PMC4417144
- DOI: 10.1093/nar/gkv056
The nucleoporin Mlp2 is involved in chromosomal distribution during mitosis in trypanosomatids
Abstract
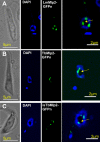
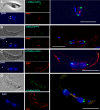
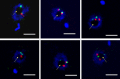
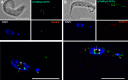
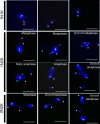
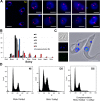
Nucleoporins are evolutionary conserved proteins mainly involved in the constitution of the nuclear pores and trafficking between the nucleus and cytoplasm, but are also increasingly viewed as main actors in chromatin dynamics and intra-nuclear mitotic events. Here, we determined the cellular localization of the nucleoporin Mlp2 in the 'divergent' eukaryotes Leishmania major and Trypanosoma brucei. In both protozoa, Mlp2 displayed an atypical localization for a nucleoporin, essentially intranuclear, and preferentially in the periphery of the nucleolus during interphase; moreover, it relocated at the mitotic spindle poles during mitosis. In T. brucei, where most centromeres have been identified, TbMlp2 was found adjacent to the centromeric sequences, as well as to a recently described unconventional kinetochore protein, in the periphery of the nucleolus, during interphase and from the end of anaphase onwards. TbMlp2 and the centromeres/kinetochores exhibited a differential migration towards the poles during mitosis. RNAi knockdown of TbMlp2 disrupted the mitotic distribution of chromosomes, leading to a surprisingly well-tolerated aneuploidy. In addition, diploidy was restored in a complementation assay where LmMlp2, the orthologue of TbMlp2 in Leishmania, was expressed in TbMlp2-RNAi-knockdown parasites. Taken together, our results demonstrate that Mlp2 is involved in the distribution of chromosomes during mitosis in trypanosomatids.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Davis L.I. The nuclear pore complex. Annu. Rev. Biochem. 1995;64:865–896. - PubMed
-
- Pante N., Aebi U. Molecular dissection of the nuclear pore complex. Crit. Rev. Biochem. Mol. Biol. 1996;31:153–199. - PubMed
-
- Gorlich D., Mattaj I.W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. - PubMed
-
- Nigg E.A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

