Role of common and rare variants in SCN10A: results from the Brugada syndrome QRS locus gene discovery collaborative study
- PMID: 25691538
- PMCID: PMC4447806
- DOI: 10.1093/cvr/cvv042
Role of common and rare variants in SCN10A: results from the Brugada syndrome QRS locus gene discovery collaborative study
Erratum in
-
Corrigendum to: Role of common and rare variants in SCN10A: results from the Brugada syndrome QRS locus gene discovery collaborative study.Cardiovasc Res. 2016 May 1;110(1):3. doi: 10.1093/cvr/cvw038. Epub 2016 Feb 24. Cardiovasc Res. 2016. PMID: 26917719 Free PMC article. No abstract available.
Abstract
Aims: Brugada syndrome (BrS) remains genetically heterogeneous and is associated with slowed cardiac conduction. We aimed to identify genetic variation in BrS cases at loci associated with QRS duration.
Methods and results: A multi-centre study sequenced seven candidate genes (SCN10A, HAND1, PLN, CASQ2, TKT, TBX3, and TBX5) in 156 Caucasian SCN5A mutation-negative BrS patients (80% male; mean age 48) with symptoms (64%) and/or a family history of sudden death (47%) or BrS (18%). Forty-nine variants were identified: 18 were rare (MAF <1%) and non-synonymous; and 11/18 (61.1%), mostly in SCN10A, were predicted as pathogenic using multiple bioinformatics tools. Allele frequencies were compared with the Exome Sequencing and UK10K Projects. SKAT methods tested rare variation in SCN10A finding no statistically significant difference between cases and controls. Co-segregation analysis was possible for four of seven probands carrying a novel pathogenic variant. Only one pedigree (I671V/G1299A in SCN10A) showed co-segregation. The SCN10A SNP V1073 was, however, associated strongly with BrS [66.9 vs. 40.1% (UK10K) OR (95% CI) = 3.02 (2.35-3.87), P = 8.07 × 10-19]. Voltage-clamp experiments for NaV1.8 were performed for SCN10A common variants V1073, A1073, and rare variants of interest: A200V and I671V. V1073, A200V and I671V, demonstrated significant reductions in peak INa compared with ancestral allele A1073 (rs6795970).
Conclusion: Rare variants in the screened QRS-associated genes (including SCN10A) are not responsible for a significant proportion of SCN5A mutation negative BrS. The common SNP SCN10A V1073 was strongly associated with BrS and demonstrated loss of NaV1.8 function, as did rare variants in isolated patients.
Keywords: Brugada syndrome; Genetics; QRS duration; Rare variants; SCN10A.
Published on behalf of the European Society of Cardiology. All rights reserved. © The Author 2015. For permissions please email: journals.permissions@oup.com.
Figures
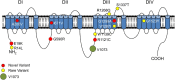



Comment in
-
Genetic background of Brugada syndrome is more complex than what we would like it to be!Cardiovasc Res. 2015 Jun 1;106(3):351-2. doi: 10.1093/cvr/cvv135. Epub 2015 Apr 29. Cardiovasc Res. 2015. PMID: 25926434 No abstract available.
References
-
- Priori SG, Wilde AA, Horie M, Cho Y, Behr ER, Berul C, Blom N, Brugada J, Chiang C-E, Huikuri H, Kannankeril P, Krahn A, Leenhardt A, Moss A, Schwartz PJ, Shimizu W, Tomaselli G, Tracy C. Executive summary: HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes. Heart Rhythm 2013;10:e85–108. - PubMed
-
- Wilde AAM, Behr ER. Genetic testing for inherited cardiac disease. Nat Rev Cardiol 2013;10:571–583. - PubMed
-
- Bezzina CR, Barc J, Mizusawa Y, Remme CA, Gourraud JB, Simonet F, Verkerk AO, Schwartz PJ, Crotti L, Dagradi F, Guicheney P, Fressart V, Leenhardt A, Antzelevitch C, Bartkowiak S, Borggrefe M, Schimpf R, Schulze-Bahr E, Zumhagen S, Behr ER, Bastiaenen R, Tfelt-Hansen J, Olesen MS, Kääb S, Beckmann BM, Weeke P, Watanabe H, Endo N, Minamino T, Horie M, Ohno S, Hasegawa K, Makita N, Nogami A, Shimizu W, Aiba T, Froguel P, Balkau B, Lantieri O, Torchio M, Wiese C, Weber D, Wolswinkel R, Coronel R, Boukens BJ, Bézieau S, Charpentier E, Chatel S, Despres A, Gros F, Kyndt F, Lecointe S, Lindenbaum P, Portero V, Violleau J, Gessler M, Tan HL, Roden DM, Christoffels VM, Le Marec H, Wilde AA, Probst V, Schott JJ, Dina C, Redon R. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet 2013;45:1044–1049. - PMC - PubMed
-
- Sotoodehnia N, Isaacs A, de Bakker PI, Dörr M, Newton-Cheh C, Nolte IM, van der Harst P, Müller M, Eijgelsheim M, Alonso A, Hicks AA, Padmanabhan S, Hayward C, Smith AV, Polasek O, Giovannone S, Fu J, Magnani JW, Marciante KD, Pfeufer A, Gharib SA, Teumer A, Li M, Bis JC, Rivadeneira F, Aspelund T, Köttgen A, Johnson T, Rice K, Sie MP, Wang YA, Klopp N, Fuchsberger C, Wild SH, Mateo Leach I, Estrada K, Völker U, Wright AF, Asselbergs FW, Qu J, Chakravarti A, Sinner MF, Kors JA, Petersmann A, Harris TB, Soliman EZ, Munroe PB, Psaty BM, Oostra BA, Cupples LA, Perz S, de Boer RA, Uitterlinden AG, Völzke H, Spector TD, Liu FY, Boerwinkle E, Dominiczak AF, Rotter JI, van Herpen G, Levy D, Wichmann HE, van Gilst WH, Witteman JC, Kroemer HK, Kao WH, Heckbert SR, Meitinger T, Hofman A, Campbell H, Folsom AR, van Veldhuisen DJ, Schwienbacher C, O'Donnell CJ, Volpato CB, Caulfield MJ, Connell JM, Launer L, Lu X, Franke L, Fehrmann RS, te Meerman G, Groen HJ, Weersma RK, van den Berg LH, Wijmenga C, Ophoff RA, Navis G, Rudan I, Snieder H, Wilson JF, Pramstaller PP, Siscovick DS, Wang TJ, Gudnason V, van Duijn CM, Felix SB, Fishman GI, Jamshidi Y, Stricker BH, Samani NJ, Kääb S, Arking DE. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet 2010;42:1068–1076. - PMC - PubMed
-
- Grantham R. Amino acid difference formula to help explain protein evolution. Science 1974;185:862–864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

