The roles of post-translational modifications in the context of protein interaction networks
- PMID: 25692714
- PMCID: PMC4333291
- DOI: 10.1371/journal.pcbi.1004049
The roles of post-translational modifications in the context of protein interaction networks
Abstract
Among other effects, post-translational modifications (PTMs) have been shown to exert their function via the modulation of protein-protein interactions. For twelve different main PTM-types and associated subtypes and across 9 diverse species, we investigated whether particular PTM-types are associated with proteins with specific and possibly "strategic" placements in the network of all protein interactions by determining informative network-theoretic properties. Proteins undergoing a PTM were observed to engage in more interactions and positioned in more central locations than non-PTM proteins. Among the twelve considered PTM-types, phosphorylated proteins were identified most consistently as being situated in central network locations and with the broadest interaction spectrum to proteins carrying other PTM-types, while glycosylated proteins are preferentially located at the network periphery. For the human interactome, proteins undergoing sumoylation or proteolytic cleavage were found with the most characteristic network properties. PTM-type-specific protein interaction network (PIN) properties can be rationalized with regard to the function of the respective PTM-carrying proteins. For example, glycosylation sites were found enriched in proteins with plasma membrane localizations and transporter or receptor activity, which generally have fewer interacting partners. The involvement in disease processes of human proteins undergoing PTMs was also found associated with characteristic PIN properties. By integrating global protein interaction networks and specific PTMs, our study offers a novel approach to unraveling the role of PTMs in cellular processes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

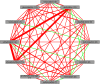
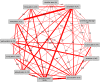


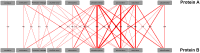
References
-
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, et al. (2000) Molecular cell biology.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

