DYn-2 Based Identification of Arabidopsis Sulfenomes
- PMID: 25693797
- PMCID: PMC4424392
- DOI: 10.1074/mcp.M114.046896
DYn-2 Based Identification of Arabidopsis Sulfenomes
Abstract
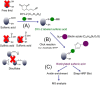
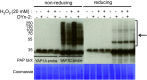
Identifying the sulfenylation state of stressed cells is emerging as a strategic approach for the detection of key reactive oxygen species signaling proteins. Here, we optimized an in vivo trapping method for cysteine sulfenic acids in hydrogen peroxide (H2O2) stressed plant cells using a dimedone based DYn-2 probe. We demonstrated that DYn-2 specifically detects sulfenylation events in an H2O2 dose- and time-dependent way. With mass spectrometry, we identified 226 sulfenylated proteins after H2O2 treatment of Arabidopsis cells, residing in the cytoplasm (123); plastid (68); mitochondria (14); nucleus (10); endoplasmic reticulum, Golgi and plasma membrane (7) and peroxisomes (4). Of these, 123 sulfenylated proteins have never been reported before to undergo cysteine oxidative post-translational modifications in plants. All in all, with this DYn-2 approach, we have identified new sulfenylated proteins, and gave a first glance on the locations of the sulfenomes of Arabidopsis thaliana.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Di Simplicio P., Franconi F., Frosalí S., Di Giuseppe D. (2003) Thiolation and nitrosation of cysteines in biological fluids and cells. Amino Acids 25, 323–339 - PubMed
-
- Jacques S., Ghesquière B., Van Breusegem F., Gevaert K. (2013) Plant proteins under oxidative attack. Proteomics 13, 932–940 - PubMed
-
- Delaunay A., Pflieger D., Barrault M.-B., Vinh J., Toledano M. B. (2002) A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111, 471–481 - PubMed
-
- Tachibana T., Okazaki S., Murayama A., Naganuma A., Nomoto A., Kuge S. (2009) A major peroxiredoxin-induced activation of Yap1 transcription factor is mediated by reduction-sensitive disulfide bonds and reveals a low level of transcriptional activation. J. Biol. Chem. 284, 4464–4472 - PubMed
-
- Chiang S. M., Schellhorn H. E. (2012) Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 525, 161–169 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

