Nanomicroarray and multiplex next-generation sequencing for simultaneous identification and characterization of influenza viruses
- PMID: 25694248
- PMCID: PMC4344273
- DOI: 10.3201/eid2103.141169
Nanomicroarray and multiplex next-generation sequencing for simultaneous identification and characterization of influenza viruses
Abstract
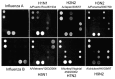
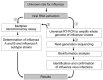
Conventional methods for detection and discrimination of influenza viruses are time consuming and labor intensive. We developed a diagnostic platform for simultaneous identification and characterization of influenza viruses that uses a combination of nanomicroarray for screening and multiplex next-generation sequencing (NGS) assays for laboratory confirmation. The nanomicroarray was developed to target hemagglutinin, neuraminidase, and matrix genes to identify influenza A and B viruses. PCR amplicons synthesized by using an adapted universal primer for all 8 gene segments of 9 influenza A subtypes were detected in the nanomicroarray and confirmed by the NGS assays. This platform can simultaneously detect and differentiate multiple influenza A subtypes in a single sample. Use of these methods as part of a new diagnostic algorithm for detection and confirmation of influenza infections may provide ongoing public health benefits by assisting with future epidemiologic studies and improving preparedness for potential influenza pandemics.
Figures


Similar articles
-
Hemagglutinin and neuraminidase genes of influenza B viruses circulating in Riyadh, Saudi Arabia during 2010-2011: evolution and sequence analysis.J Med Virol. 2014 Jun;86(6):1003-16. doi: 10.1002/jmv.23819. Epub 2013 Oct 22. J Med Virol. 2014. PMID: 24150926
-
Multiplex Reverse Transcription-PCR for Simultaneous Surveillance of Influenza A and B Viruses.J Clin Microbiol. 2017 Dec;55(12):3492-3501. doi: 10.1128/JCM.00957-17. Epub 2017 Oct 4. J Clin Microbiol. 2017. PMID: 28978683 Free PMC article.
-
Multiplex RT-PCR and indirect immunofluorescence assays for detection and subtyping of human influenza virus in Tunisia.Curr Microbiol. 2015 Mar;70(3):324-9. doi: 10.1007/s00284-014-0719-0. Epub 2014 Nov 1. Curr Microbiol. 2015. PMID: 25366277
-
Influenza virus: Genomic insights, evolution, and its clinical presentation.Microb Pathog. 2025 Aug;205:107671. doi: 10.1016/j.micpath.2025.107671. Epub 2025 May 7. Microb Pathog. 2025. PMID: 40345348 Review.
-
Current Approaches for Diagnosis of Influenza Virus Infections in Humans.Viruses. 2016 Apr 12;8(4):96. doi: 10.3390/v8040096. Viruses. 2016. PMID: 27077877 Free PMC article. Review.
Cited by
-
Graphene functionalized field-effect transistors for ultrasensitive detection of Japanese encephalitis and Avian influenza virus.Sci Rep. 2020 Sep 3;10(1):14546. doi: 10.1038/s41598-020-71591-w. Sci Rep. 2020. PMID: 32884083 Free PMC article.
-
Direct RNA Sequencing of the Coding Complete Influenza A Virus Genome.Sci Rep. 2018 Sep 26;8(1):14408. doi: 10.1038/s41598-018-32615-8. Sci Rep. 2018. PMID: 30258076 Free PMC article.
-
Sensitive Detection and Simultaneous Discrimination of Influenza A and B Viruses in Nasopharyngeal Swabs in a Single Assay Using Next-Generation Sequencing-Based Diagnostics.PLoS One. 2016 Sep 22;11(9):e0163175. doi: 10.1371/journal.pone.0163175. eCollection 2016. PLoS One. 2016. PMID: 27658193 Free PMC article.
-
Advances in laboratory assays for detecting human metapneumovirus.Ann Transl Med. 2020 May;8(9):608. doi: 10.21037/atm.2019.12.42. Ann Transl Med. 2020. PMID: 32566634 Free PMC article. Review.
-
Universal primers for rift valley fever virus whole-genome sequencing.Sci Rep. 2023 Oct 31;13(1):18688. doi: 10.1038/s41598-023-45848-z. Sci Rep. 2023. PMID: 37907670 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

