The transcription factor Mesp1 interacts with cAMP-responsive element binding protein 1 (Creb1) and coactivates Ets variant 2 (Etv2) gene expression
- PMID: 25694434
- PMCID: PMC4392264
- DOI: 10.1074/jbc.M114.614628
The transcription factor Mesp1 interacts with cAMP-responsive element binding protein 1 (Creb1) and coactivates Ets variant 2 (Etv2) gene expression
Abstract
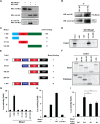
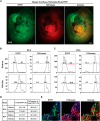
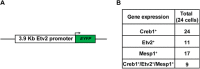
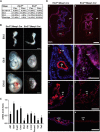
Mesoderm posterior 1 (Mesp1) is well recognized for its role in cardiac development, although it is expressed broadly in mesodermal lineages. We have previously demonstrated important roles for Mesp1 and Ets variant 2 (Etv2) during lineage specification, but their relationship has not been defined. This study reveals that Mesp1 binds to the proximal promoter and transactivates Etv2 gene expression via the CRE motif. We also demonstrate the protein-protein interaction between Mesp1 and cAMP-responsive element binding protein 1 (Creb1) in vitro and in vivo. Utilizing transgenesis, lineage tracing, flow cytometry, and immunostaining technologies, we define the lineage relationship between Mesp1- and Etv2-expressing cell populations. We observe that the majority of Etv2-EYFP(+) cells are derived from Mesp1-Cre(+) cells in both the embryo and yolk sac. Furthermore, we observe that the conditional deletion of Etv2, using a Mesp1-Cre transgenic strategy, results in vascular and hematopoietic defects similar to those observed in the global deletion of Etv2 and that it has embryonic lethality by embryonic day 9.5. In summary, our study supports the hypothesis that Mesp1 is a direct upstream transactivator of Etv2 during embryogenesis and that Creb1 is an important cofactor of Mesp1 in the transcriptional regulation of Etv2 gene expression.
Keywords: Basic Helix-Loop-Helix Transcription Factor (bHLH); ETS Transcription Factor Family; Protein-Protein Interaction; Transcription Regulation; Transcriptional Coactivator.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Keller G., Lacaud G., Robertson S. (1999) Development of the hematopoietic system in the mouse. Exp. Hematol. 27, 777–787 - PubMed
-
- Ferkowicz M. J., Yoder M. C. (2005) Blood island formation: longstanding observations and modern interpretations. Exp. Hematol. 33, 1041–1047 - PubMed
-
- Li W., Ferkowicz M. J., Johnson S. A., Shelley W. C., Yoder M. C. (2005) Endothelial cells in the early murine yolk sac give rise to CD41-expressing hematopoietic cells. Stem. Cells Dev. 14, 44–54 - PubMed
-
- Wilson N. K., Foster S. D., Wang X., Knezevic K., Schütte J., Kaimakis P., Chilarska P. M., Kinston S., Ouwehand W. H., Dzierzak E., Pimanda J. E., de Bruijn M. F., Göttgens B. (2010) Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7, 532–544 - PubMed
-
- Moignard V., Woodhouse S., Fisher J., Göttgens B. (2013) Transcriptional hierarchies regulating early blood cell development. Blood Cells Mol. Dis. 51, 239–247 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

