Adenovirus replaces mitotic checkpoint controls
- PMID: 25694601
- PMCID: PMC4403466
- DOI: 10.1128/JVI.00213-15
Adenovirus replaces mitotic checkpoint controls
Abstract
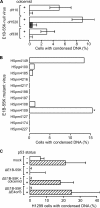
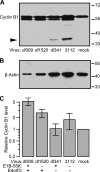
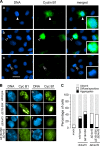
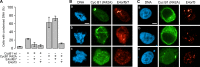
Infection with adenovirus triggers the cellular DNA damage response, elements of which include cell death and cell cycle arrest. Early adenoviral proteins, including the E1B-55K and E4orf3 proteins, inhibit signaling in response to DNA damage. A fraction of cells infected with an adenovirus mutant unable to express the E1B-55K and E4orf3 genes appeared to arrest in a mitotic-like state. Cells infected early in G1 of the cell cycle were predisposed to arrest in this state at late times of infection. This arrested state, which displays hallmarks of mitotic catastrophe, was prevented by expression of either the E1B-55K or the E4orf3 genes. However, E1B-55K mutant virus-infected cells became trapped in a mitotic-like state in the presence of the microtubule poison colcemid, suggesting that the two viral proteins restrict entry into mitosis or facilitate exit from mitosis in order to prevent infected cells from arresting in mitosis. The E1B-55K protein appeared to prevent inappropriate entry into mitosis through its interaction with the cellular tumor suppressor protein p53. The E4orf3 protein facilitated exit from mitosis by possibly mislocalizing and functionally inactivating cyclin B1. When expressed in noninfected cells, E4orf3 overcame the mitotic arrest caused by the degradation-resistant R42A cyclin B1 variant.
Importance: Cells that are infected with adenovirus type 5 early in G1 of the cell cycle are predisposed to arrest in a mitotic-like state in a p53-dependent manner. The adenoviral E1B-55K protein prevents entry into mitosis. This newly described activity for the E1B-55K protein appears to depend on the interaction between the E1B-55K protein and the tumor suppressor p53. The adenoviral E4orf3 protein facilitates exit from mitosis, possibly by altering the intracellular distribution of cyclin B1. By preventing entry into mitosis and by promoting exit from mitosis, these adenoviral proteins act to prevent the infected cell from arresting in a mitotic-like state.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
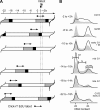
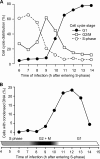
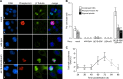
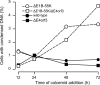
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

