A complex genome-microRNA interplay in human mitochondria
- PMID: 25695052
- PMCID: PMC4324738
- DOI: 10.1155/2015/206382
A complex genome-microRNA interplay in human mitochondria
Abstract
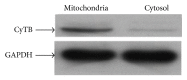
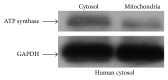
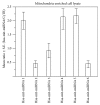
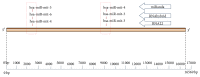
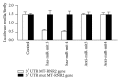
Small noncoding regulatory RNA exist in wide spectrum of organisms ranging from prokaryote bacteria to humans. In human, a systematic search for noncoding RNA is mainly limited to the nuclear and cytosolic compartments. To investigate whether endogenous small regulatory RNA are present in cell organelles, human mitochondrial genome was also explored for prediction of precursor microRNA (pre-miRNA) and mature miRNA (miRNA) sequences. Six novel miRNA were predicted from the organelle genome by bioinformatics analysis. The structures are conserved in other five mammals including chimp, orangutan, mouse, rat, and rhesus genome. Experimentally, six human miRNA are well accumulated or deposited in human mitochondria. Three of them are expressed less prominently in Northern analysis. To ascertain their presence in human skeletal muscles, total RNA was extracted from enriched mitochondria by an immunomagnetic method. The expression of six novel pre-miRNA and miRNA was confirmed by Northern blot analysis; however, low level of remaining miRNA was found by sensitive Northern analysis. Their presence is further confirmed by real time RT-PCR. The six miRNA find their multiple targets throughout the human genome in three different types of software. The luciferase assay was used to confirm that MT-RNR2 gene was the potential target of hsa-miR-mit3 and hsa-miR-mit4.
Figures









References
-
- Brown M. D., Wallace D. C. Spectrum of mitochondrial DNA mutations in Leber's hereditary optic neuropathy. Clinical Neuroscience. 1994:138–145.
-
- Easteal S. The relative rate of DNA evolution in primates. Molecular Biology and Evolution. 1991;8(1):115–127. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

