Regulation of IGFBP-2 expression during fasting
- PMID: 25695641
- PMCID: PMC4403943
- DOI: 10.1042/BJ20141248
Regulation of IGFBP-2 expression during fasting
Abstract
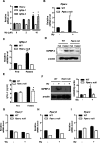
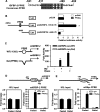
Insulin-like growth factor (IGF)-binding protein-2 (IGFBP-2), one of the most abundant circulating IGFBPs, is known to attenuate the biological action of IGF-1. Although the effect of IGFBP-2 in preventing metabolic disorders is well known, its regulatory mechanism remains unclear. In the present study, we demonstrated the transcriptional regulation of the Igfbp-2 gene by peroxisome-proliferator-activated receptor (PPAR) α in the liver. During fasting, both Igfbp-2 and PPARα expression levels were increased. Wy14643, a selective PPARα agonist, significantly induced Igfbp-2 gene expression in primary cultured hepatocytes. However, Igfbp-2 gene expression in Pparα null mice was not affected by fasting or Wy14643. In addition, through transient transfection and chromatin immunoprecipitation assay in fasted livers, we determined that PPARα bound to the putative PPAR-responsive element between -511 bp and -499 bp on the Igfbp-2 gene promoter, indicating that the Igfbp-2 gene transcription is activated directly by PPARα. To explore the role of PPARα in IGF-1 signalling, we treated primary cultured hepatocytes with Wy14643 and observed a decrease in the number of IGF-1 receptors (IGF-1Rs) and in Akt phosphorylation. No inhibition was observed in the hepatocytes isolated from Pparα null mice. These results suggest that PPARα controls IGF-1 signalling through the up-regulation of hepatic Igfbp-2 transcription during fasting and Wy14643 treatment.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

