Data-driven prediction and design of bZIP coiled-coil interactions
- PMID: 25695764
- PMCID: PMC4335062
- DOI: 10.1371/journal.pcbi.1004046
Data-driven prediction and design of bZIP coiled-coil interactions
Erratum in
-
Correction: Data-driven prediction and design of bZIP coiled-coil interactions.PLoS Comput Biol. 2015 Apr 15;11(4):e1004243. doi: 10.1371/journal.pcbi.1004243. eCollection 2015 Apr. PLoS Comput Biol. 2015. PMID: 25874775 Free PMC article. No abstract available.
Abstract
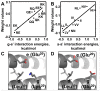
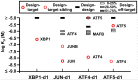
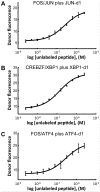
Selective dimerization of the basic-region leucine-zipper (bZIP) transcription factors presents a vivid example of how a high degree of interaction specificity can be achieved within a family of structurally similar proteins. The coiled-coil motif that mediates homo- or hetero-dimerization of the bZIP proteins has been intensively studied, and a variety of methods have been proposed to predict these interactions from sequence data. In this work, we used a large quantitative set of 4,549 bZIP coiled-coil interactions to develop a predictive model that exploits knowledge of structurally conserved residue-residue interactions in the coiled-coil motif. Our model, which expresses interaction energies as a sum of interpretable residue-pair and triplet terms, achieves a correlation with experimental binding free energies of R = 0.68 and significantly out-performs other scoring functions. To use our model in protein design applications, we devised a strategy in which synthetic peptides are built by assembling 7-residue native-protein heptad modules into new combinations. An integer linear program was used to find the optimal combination of heptads to bind selectively to a target human bZIP coiled coil, but not to target paralogs. Using this approach, we designed peptides to interact with the bZIP domains from human JUN, XBP1, ATF4 and ATF5. Testing more than 132 candidate protein complexes using a fluorescence resonance energy transfer assay confirmed the formation of tight and selective heterodimers between the designed peptides and their targets. This approach can be used to make inhibitors of native proteins, or to develop novel peptides for applications in synthetic biology or nanotechnology.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Increasing the affinity of selective bZIP-binding peptides through surface residue redesign.Protein Sci. 2014 Jul;23(7):940-53. doi: 10.1002/pro.2477. Epub 2014 Apr 30. Protein Sci. 2014. PMID: 24729132 Free PMC article.
-
Identification of bZIP interaction partners of viral proteins HBZ, MEQ, BZLF1, and K-bZIP using coiled-coil arrays.Biochemistry. 2010 Mar 9;49(9):1985-97. doi: 10.1021/bi902065k. Biochemistry. 2010. PMID: 20102225 Free PMC article.
-
Structure-based prediction of bZIP partnering specificity.J Mol Biol. 2006 Feb 3;355(5):1125-42. doi: 10.1016/j.jmb.2005.11.036. Epub 2005 Dec 1. J Mol Biol. 2006. PMID: 16359704
-
Pharmacological interference with protein-protein interactions mediated by coiled-coil motifs.Handb Exp Pharmacol. 2008;(186):461-82. doi: 10.1007/978-3-540-72843-6_19. Handb Exp Pharmacol. 2008. PMID: 18491064 Review.
-
Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: A molecular model of a wheat bZIP factor and implications of its structure in function.Biochim Biophys Acta. 2016 Jan;1860(1 Pt A):46-56. doi: 10.1016/j.bbagen.2015.10.014. Epub 2015 Oct 20. Biochim Biophys Acta. 2016. PMID: 26493723 Review.
Cited by
-
Correction: Data-driven prediction and design of bZIP coiled-coil interactions.PLoS Comput Biol. 2015 Apr 15;11(4):e1004243. doi: 10.1371/journal.pcbi.1004243. eCollection 2015 Apr. PLoS Comput Biol. 2015. PMID: 25874775 Free PMC article. No abstract available.
-
A Barcoding Strategy Enabling Higher-Throughput Library Screening by Microscopy.ACS Synth Biol. 2015 Nov 20;4(11):1205-16. doi: 10.1021/acssynbio.5b00060. Epub 2015 Jul 15. ACS Synth Biol. 2015. PMID: 26155738 Free PMC article.
-
Molecular mechanisms of the protein-protein interaction-regulated binding specificity of basic-region leucine zipper transcription factors.J Mol Model. 2019 Jul 24;25(8):246. doi: 10.1007/s00894-019-4138-9. J Mol Model. 2019. PMID: 31342181
-
Prediction of Protein-Protein Binding Interactions in Dimeric Coiled Coils by Information Contained in Folding Energy Landscapes.Int J Mol Sci. 2021 Jan 29;22(3):1368. doi: 10.3390/ijms22031368. Int J Mol Sci. 2021. PMID: 33573048 Free PMC article.
-
A general-purpose protein design framework based on mining sequence-structure relationships in known protein structures.Proc Natl Acad Sci U S A. 2020 Jan 14;117(2):1059-1068. doi: 10.1073/pnas.1908723117. Epub 2019 Dec 31. Proc Natl Acad Sci U S A. 2020. PMID: 31892539 Free PMC article.
References
-
- Newman JRS, Keating AE (2003) Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300: 2097–2101. - PubMed
-
- O’Neil KT, DeGrado WF (1990) A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science 250: 646–651. - PubMed
-
- Harbury PB, Zhang T, Kim PS, Alber T (1993) A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262: 1401–1407. - PubMed
-
- Krylov D, Barchi J, Vinson C (1998) Inter-helical interactions in the leucine zipper coiled coil dimer: pH and salt dependence of coupling energy between charged amino acids. J Mol Biol 279: 959–972. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

