Molecular mechanism of HIV-1 Tat interacting with human dopamine transporter
- PMID: 25695767
- PMCID: PMC4400243
- DOI: 10.1021/acschemneuro.5b00001
Molecular mechanism of HIV-1 Tat interacting with human dopamine transporter
Abstract
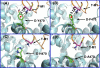
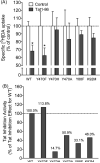
Nearly 70% of HIV-1-infected individuals suffer from HIV-associated neurocognitive disorders (HAND). HIV-1 transactivator of transcription (Tat) protein is known to synergize with abused drugs and exacerbate the progression of central nervous system (CNS) pathology. Cumulative evidence suggest that the HIV-1 Tat protein exerts the neurotoxicity through interaction with human dopamine transporter (hDAT) in the CNS. Through computational modeling and molecular dynamics (MD) simulations, we develop a three-dimensional (3D) structural model for HIV-1 Tat binding with hDAT. The model provides novel mechanistic insights concerning how HIV-1 Tat interacts with hDAT and inhibits dopamine uptake by hDAT. In particular, according to the computational modeling, Tat binds most favorably with the outward-open state of hDAT. Residues Y88, K92, and Y470 of hDAT are predicted to be key residues involved in the interaction between hDAT and Tat. The roles of these hDAT residues in the interaction with Tat are validated by experimental tests through site-directed mutagensis and dopamine uptake assays. The agreement between the computational and experimental data suggests that the computationally predicted hDAT-Tat binding mode and mechanistic insights are reasonable and provide a new starting point to design further pharmacological studies on the molecular mechanism of HIV-1-associated neurocognitive disorders.
Keywords: Transactivator of transcription; dopamine uptake; neurotoxicity; protein−protein interaction; viral protein.
Figures

Similar articles
-
Mutations of Human DopamineTransporter at Tyrosine88, Aspartic Acid206, and Histidine547 Influence Basal and HIV-1 Tat-inhibited Dopamine Transport.J Neuroimmune Pharmacol. 2021 Dec;16(4):854-869. doi: 10.1007/s11481-021-09984-5. Epub 2021 Feb 3. J Neuroimmune Pharmacol. 2021. PMID: 33537927 Free PMC article.
-
Role of Histidine 547 of Human Dopamine Transporter in Molecular Interaction with HIV-1 Tat and Dopamine Uptake.Sci Rep. 2016 Jun 2;6:27314. doi: 10.1038/srep27314. Sci Rep. 2016. PMID: 27250920 Free PMC article.
-
Molecular mechanism: the human dopamine transporter histidine 547 regulates basal and HIV-1 Tat protein-inhibited dopamine transport.Sci Rep. 2016 Dec 14;6:39048. doi: 10.1038/srep39048. Sci Rep. 2016. PMID: 27966610 Free PMC article.
-
The role of human dopamine transporter in NeuroAIDS.Pharmacol Ther. 2018 Mar;183:78-89. doi: 10.1016/j.pharmthera.2017.10.007. Epub 2017 Oct 5. Pharmacol Ther. 2018. PMID: 28987321 Free PMC article. Review.
-
HIV, Tat and dopamine transmission.Neurobiol Dis. 2017 Sep;105:51-73. doi: 10.1016/j.nbd.2017.04.015. Epub 2017 Apr 27. Neurobiol Dis. 2017. PMID: 28457951 Free PMC article. Review.
Cited by
-
Binding Mode of Human Norepinephrine Transporter Interacting with HIV-1 Tat.ACS Chem Neurosci. 2021 May 5;12(9):1519-1527. doi: 10.1021/acschemneuro.0c00792. Epub 2021 Apr 22. ACS Chem Neurosci. 2021. PMID: 33886267 Free PMC article.
-
Neurological, Behavioral, and Pathophysiological Characterization of the Co-Occurrence of Substance Use and HIV: A Narrative Review.Brain Sci. 2023 Oct 19;13(10):1480. doi: 10.3390/brainsci13101480. Brain Sci. 2023. PMID: 37891847 Free PMC article. Review.
-
Structure-Based Design and Discovery of a Long-Acting Cocaine Hydrolase Mutant with Improved Binding Affinity to Neonatal Fc Receptor for Treatment of Cocaine Abuse.AAPS J. 2020 Mar 18;22(3):62. doi: 10.1208/s12248-020-00442-3. AAPS J. 2020. PMID: 32189158 Free PMC article.
-
Suppression of HIV and cocaine-induced neurotoxicity and inflammation by cell penetrable itaconate esters.bioRxiv [Preprint]. 2023 Sep 26:2023.09.25.559154. doi: 10.1101/2023.09.25.559154. bioRxiv. 2023. Update in: J Neurovirol. 2024 Aug;30(4):337-352. doi: 10.1007/s13365-024-01216-9. PMID: 37808776 Free PMC article. Updated. Preprint.
-
Mutations of Human DopamineTransporter at Tyrosine88, Aspartic Acid206, and Histidine547 Influence Basal and HIV-1 Tat-inhibited Dopamine Transport.J Neuroimmune Pharmacol. 2021 Dec;16(4):854-869. doi: 10.1007/s11481-021-09984-5. Epub 2021 Feb 3. J Neuroimmune Pharmacol. 2021. PMID: 33537927 Free PMC article.
References
-
- Ferris MJ, Frederick-Duus D, Fadel J, Mactutus CF, Booze RM. The human immunodeficiency virus-1–associated protein, Tat1-86, impairs dopamine transporters and interacts with cocaine to reduce nerve terminal function: A no-net-flux microdialysis study. Neuroscience. 2009;159:1292–1299. - PMC - PubMed
-
- Valdiserri RO. Commentary: Thirty Years of AIDS in America: A Story of Infinite Hope. AIDS Education and Prevention. 2011;23:479–494. - PubMed
-
- Who, U., and Unicef . Global report: UNAIDS report on the global AIDS epidemic 2013. UNAIDS; Geneva: 2013.
-
- Wallace DR, Dodson S, Nath A, Booze RM. Estrogen attenuates gp120-and tat1–72-induced oxidative stress and prevents loss of dopamine transporter function. Synapse. 2006;59:51–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

