Rewired metabolism in drug-resistant leukemia cells: a metabolic switch hallmarked by reduced dependence on exogenous glutamine
- PMID: 25697355
- PMCID: PMC4375488
- DOI: 10.1074/jbc.M114.618769
Rewired metabolism in drug-resistant leukemia cells: a metabolic switch hallmarked by reduced dependence on exogenous glutamine
Abstract
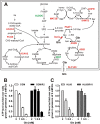
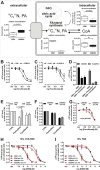
Cancer cells that escape induction therapy are a major cause of relapse. Understanding metabolic alterations associated with drug resistance opens up unexplored opportunities for the development of new therapeutic strategies. Here, we applied a broad spectrum of technologies including RNA sequencing, global untargeted metabolomics, and stable isotope labeling mass spectrometry to identify metabolic changes in P-glycoprotein overexpressing T-cell acute lymphoblastic leukemia (ALL) cells, which escaped a therapeutically relevant daunorubicin treatment. We show that compared with sensitive ALL cells, resistant leukemia cells possess a fundamentally rewired central metabolism characterized by reduced dependence on glutamine despite a lack of expression of glutamate-ammonia ligase (GLUL), a higher demand for glucose and an altered rate of fatty acid β-oxidation, accompanied by a decreased pantothenic acid uptake capacity. We experimentally validate our findings by selectively targeting components of this metabolic switch, using approved drugs and starvation approaches followed by cell viability analyses in both the ALL cells and in an acute myeloid leukemia (AML) sensitive/resistant cell line pair. We demonstrate how comparative metabolomics and RNA expression profiling of drug-sensitive and -resistant cells expose targetable metabolic changes and potential resistance markers. Our results show that drug resistance is associated with significant metabolic costs in cancer cells, which could be exploited using new therapeutic strategies.
Keywords: Drug Resistance; Glutamine; Glycolysis; Leukemia; Mass Spectrometry (MS); Metabolism; Metabolomics; Transcriptomics; β-Oxidation.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Haffner M. C., Mosbruger T., Esopi D. M., Fedor H., Heaphy C. M., Walker D. A., Adejola N., Gürel M., Hicks J., Meeker A. K., Halushka M. K., Simons J. W., Isaacs W. B., De Marzo A. M., Nelson W. G., Yegnasubramanian S. (2013) Tracking the clonal origin of lethal prostate cancer. J. Clin. Investig. 123, 4918–4922 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

