Genome-wide DNA methylation identifies trophoblast invasion-related genes: Claudin-4 and Fucosyltransferase IV control mobility via altering matrix metalloproteinase activity
- PMID: 25697377
- PMCID: PMC4407676
- DOI: 10.1093/molehr/gav007
Genome-wide DNA methylation identifies trophoblast invasion-related genes: Claudin-4 and Fucosyltransferase IV control mobility via altering matrix metalloproteinase activity
Abstract
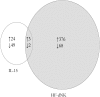
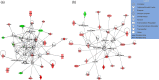
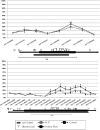
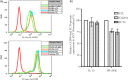
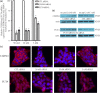
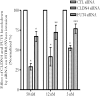
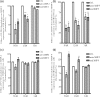
Previously we showed that extravillous cytotrophoblast (EVT) outgrowth and migration on a collagen gel explant model were affected by exposure to decidual natural killer cells (dNK). This study investigates the molecular causes behind this phenomenon. Genome wide DNA methylation of exposed and unexposed EVT was assessed using the Illumina Infinium HumanMethylation450 BeadChip array (450 K array). We identified 444 differentially methylated CpG loci in dNK-treated EVT compared with medium control (P < 0.05). The genes associated with these loci had critical biological roles in cellular development, cellular growth and proliferation, cell signaling, cellular assembly and organization by Ingenuity Pathway Analysis (IPA). Furthermore, 23 mobility-related genes were identified by IPA from dNK-treated EVT. Among these genes, CLDN4 (encoding claudin-4) and FUT4 (encoding fucosyltransferase IV) were chosen for follow-up studies because of their biological relevance from research on tumor cells. The results showed that the mRNA and protein expressions of both CLDN4 and FUT4 in dNK-treated EVT were significantly reduced compared with control (P < 0.01 for both CLDN4 and FUT4 mRNA expression; P < 0.001 for CLDN4 and P < 0.01 for FUT4 protein expression), and were inversely correlated with DNA methylation. Knocking down CLDN4 and FUT4 by small interfering RNA reduced trophoblast invasion, possibly through the altered matrix metalloproteinase (MMP)-2 and/or MMP-9 expression and activity. Taken together, dNK alter EVT mobility at least partially in association with an alteration of DNA methylation profile. Hypermethylation of CLDN4 and FUT4 reduces protein expression. CLDN4 and FUT4 are representative genes that participate in modulating trophoblast mobility.
Keywords: Claudin-4; DNA methylation; Fucosyltransferase IV; decidual natural killer cells; extravillous cytotrophoblast.
© The Author 2015. Published by Oxford University Press on behalf of the European Society of Human Reproduction and Embryology. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
References
-
- Agarwal R, D'Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res 2005;65:7378–7385. - PubMed
-
- Bertagnolo V, Benedusi M, Brugnoli F, Lanuti P, Marchisio M, Querzoli P, Capitani S. Phospholipase C-beta 2 promotes mitosis and migration of human breast cancer-derived cells. Carcinogenesis 2007;28:1638–1645. - PubMed
-
- Boireau S, Buchert M, Samuel MS, Pannequin J, Ryan JL, Choquet A, Chapuis H, Rebillard X, Avances C, Ernst M, et al. DNA-methylation-dependent alterations of claudin-4 expression in human bladder carcinoma. Carcinogenesis 2007;28:246–258. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

