The identification of new cytosolic glutamine synthetase and asparagine synthetase genes in barley (Hordeum vulgare L.), and their expression during leaf senescence
- PMID: 25697791
- PMCID: PMC4378633
- DOI: 10.1093/jxb/erv003
The identification of new cytosolic glutamine synthetase and asparagine synthetase genes in barley (Hordeum vulgare L.), and their expression during leaf senescence
Abstract
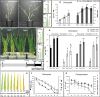
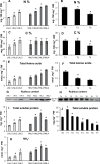
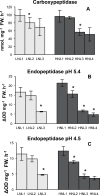
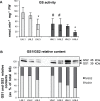
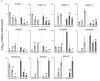
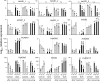
Glutamine synthetase and asparagine synthetase are two master enzymes involved in ammonium assimilation in plants. Their roles in nitrogen remobilization and nitrogen use efficiency have been proposed. In this report, the genes coding for the cytosolic glutamine synthetases (HvGS1) and asparagine synthetases (HvASN) in barley were identified. In addition to the three HvGS1 and two HvASN sequences previously reported, two prokaryotic-like HvGS1 and three HvASN cDNA sequences were identified. Gene structures were then characterized, obtaining full genomic sequences. The response of the five HvGS1 and five HvASN genes to leaf senescence was then studied. Developmental senescence was studied using primary and flag leaves. Dark-exposure or low-nitrate conditions were also used to trigger stress-induced senescence. Well-known senescence markers such as the chlorophyll and Rubisco contents were monitored in order to characterize senescence levels in the different leaves. The three eukaryotic-like HvGS1_1, HvGS1_2, and HvGS1_3 sequences showed the typical senescence-induced reduction in gene expression described in many plant species. By contrast, the two prokaryotic-like HvGS1_4 and HvGS1_5 sequences were repressed by leaf senescence, similar to the HvGS2 gene, which encodes the chloroplast glutamine synthetase isoenzyme. There was a greater contrast in the responses of the five HvASN and this suggested that these genes are needed for N remobilization in senescing leaves only when plants are well fertilized with nitrate. Responses of the HvASN sequences to dark-induced senescence showed that there are two categories of asparagine synthetases, one induced in the dark and the other repressed by the same conditions.
Keywords: Asparagine synthetase; dark treatment; endoprotease; glutamine synthetase; nitrate..
© The Author 2015. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures



References
-
- Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C. 2014. Autophagy, plant senescence, and nutrient recycling. Journal of Experimental Botany 65, 3799–3811. - PubMed
-
- Baima S, Haegi A, Stroman P, Casadoro G. 1989. Characterization of a cDNA clone for Barley leaf glutamine-synthetase. Carlsberg Research Communications 54, 1–9. - PubMed
-
- Brugière N, Dubois F, Masclaux C, Sangwan RS, Hirel B. 2000. Immunolocalization of glutamine synthetase in senescing tobacco (Nicotiana tabacum L.) leaves suggests that ammonia assimilation is progressively shifted to the mesophyll cytosol. Planta 211, 519–527. - PubMed
-
- Buchanan-Wollaston V. 1997. The molecular biology of leaf senescence. Journal of Experimental Botany 48, 181–199.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

