CapsidMaps: protein-protein interaction pattern discovery platform for the structural analysis of virus capsids using Google Maps
- PMID: 25697908
- PMCID: PMC4750372
- DOI: 10.1016/j.jsb.2015.02.003
CapsidMaps: protein-protein interaction pattern discovery platform for the structural analysis of virus capsids using Google Maps
Abstract
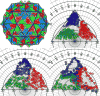
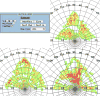
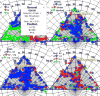
Structural analysis and visualization of protein-protein interactions is a challenging task since it is difficult to appreciate easily the extent of all contacts made by the residues forming the interfaces. In the case of viruses, structural analysis becomes even more demanding because several interfaces coexist and, in most cases, these are formed by hundreds of contacting residues that belong to multiple interacting coat proteins. CapsidMaps is an interactive analysis and visualization tool that is designed to benefit the structural virology community. Developed as an improved extension of the φ-ψ Explorer, here we describe the details of its design and implementation. We present results of analysis of a spherical virus to showcase the features and utility of the new tool. CapsidMaps also facilitates the comparison of quaternary interactions between two spherical virus particles by computing a similarity (S)-score. The tool can also be used to identify residues that are solvent exposed and in the process of locating antigenic epitope regions as well as residues forming the inside surface of the capsid that interact with the nucleic acid genome. CapsidMaps is part of the VIPERdb Science Gateway, and is freely available as a web-based and cross-browser compliant application at http://viperdb.scripps.edu.
Keywords: Heat maps; Quaternary interactions; Similarity score; Viral capsids; Web interface.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
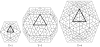



Similar articles
-
VIPERdb2: an enhanced and web API enabled relational database for structural virology.Nucleic Acids Res. 2009 Jan;37(Database issue):D436-42. doi: 10.1093/nar/gkn840. Epub 2008 Nov 3. Nucleic Acids Res. 2009. PMID: 18981051 Free PMC article.
-
VIPERdb: A Tool for Virus Research.Annu Rev Virol. 2018 Sep 29;5(1):477-488. doi: 10.1146/annurev-virology-092917-043405. Annu Rev Virol. 2018. PMID: 30265627
-
A novel method to map and compare protein-protein interactions in spherical viral capsids.Proteins. 2008 Nov 15;73(3):644-55. doi: 10.1002/prot.22088. Proteins. 2008. PMID: 18491385 Free PMC article.
-
Plant viruses as biotemplates for materials and their use in nanotechnology.Annu Rev Phytopathol. 2008;46:361-84. doi: 10.1146/annurev.phyto.032508.131939. Annu Rev Phytopathol. 2008. PMID: 18473700 Review.
-
Protein-protein interaction and quaternary structure.Q Rev Biophys. 2008 May;41(2):133-80. doi: 10.1017/S0033583508004708. Q Rev Biophys. 2008. PMID: 18812015 Review.
Cited by
-
Design Rules for the Sequestration of Viruses into Polypeptide Complex Coacervates.Biomacromolecules. 2024 Feb 12;25(2):741-753. doi: 10.1021/acs.biomac.3c00938. Epub 2023 Dec 16. Biomacromolecules. 2024. PMID: 38103178 Free PMC article.
-
A Roadmap for Building Waterborne Virus Traps.JACS Au. 2022 Oct 4;2(10):2205-2221. doi: 10.1021/jacsau.2c00377. eCollection 2022 Oct 24. JACS Au. 2022. PMID: 36311831 Free PMC article. Review.
-
Empty and Full AAV Capsid Charge and Hydrophobicity Differences Measured with Single-Particle AFM.Langmuir. 2023 Apr 25;39(16):5641-5648. doi: 10.1021/acs.langmuir.2c02643. Epub 2023 Apr 11. Langmuir. 2023. PMID: 37040364 Free PMC article.
-
Improved Virus Isoelectric Point Estimation by Exclusion of Known and Predicted Genome-Binding Regions.Appl Environ Microbiol. 2020 Nov 10;86(23):e01674-20. doi: 10.1128/AEM.01674-20. Print 2020 Nov 10. Appl Environ Microbiol. 2020. PMID: 32978129 Free PMC article.
-
Adaption of FMDV Asia-1 to Suspension Culture: Cell Resistance Is Overcome by Virus Capsid Alterations.Viruses. 2017 Aug 18;9(8):231. doi: 10.3390/v9080231. Viruses. 2017. PMID: 28820470 Free PMC article.
References
-
- Caspar Dt, Klug A. Physical principles in the construction of regular viruses. 1st. Vol. 27. Press, Cold Spring Harbor Laboratory; 1962. - PubMed
-
- Damodaran K, Reddy VS, Johnson JE, Brooks CL., III A general method to quantify quasi-equivalence in icosahedral viruses. Mol Biol. 2002;324:723–737. - PubMed
-
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of molecular biology. 2000;302(1):205–217. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

