Pseudogene-derived lncRNAs: emerging regulators of gene expression
- PMID: 25699073
- PMCID: PMC4316772
- DOI: 10.3389/fgene.2014.00476
Pseudogene-derived lncRNAs: emerging regulators of gene expression
Abstract
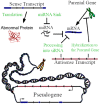
In the more than one decade since the completion of the Human Genome Project, the prevalence of non-protein-coding functional elements in the human genome has emerged as a key revelation in post-genomic biology. Highlighted by the ENCODE (Encyclopedia of DNA Elements) and FANTOM (Functional Annotation of Mammals) consortia, these elements include tens of thousands of pseudogenes, as well as comparably numerous long non-coding RNA (lncRNA) genes. Pseudogene transcription and function remain insufficiently understood. However, the field is of great importance for human disease due to the high sequence similarity between pseudogenes and their parental protein-coding genes, which generates the potential for sequence-specific regulation. Recent case studies have established essential and coordinated roles of both pseudogenes and lncRNAs in development and disease in metazoan systems, including functional impacts of lncRNA transcription at pseudogene loci on the regulation of the pseudogenes' parental genes. This review synthesizes the nascent evidence for regulatory modalities jointly exerted by lncRNAs and pseudogenes in human disease, and for recent evolutionary origins of these systems.
Keywords: genetic; genome wide; lncRNA; ncRNA; pseudogenes; regulation of gene expression; transcription.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

