Mining for viral fragments in methylation enriched sequencing data
- PMID: 25699076
- PMCID: PMC4316777
- DOI: 10.3389/fgene.2015.00016
Mining for viral fragments in methylation enriched sequencing data
Abstract
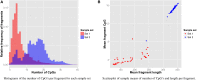
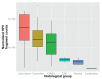
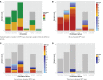
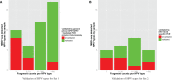
Most next generation sequencing experiments generate more data than is usable for the experimental set up. For example, methyl-CpG binding domain (MBD) affinity purification based sequencing is often used for DNA-methylation profiling, but up to 30% of the sequenced fragments cannot be mapped uniquely to the reference genome. Here we present and evaluate a methodology for the identification of viruses in these otherwise unused paired-end MBD-seq data. Viral detection is accomplished by mapping non-reference alignable reads to a comprehensive set of viral genomes. As viruses play an important role in epigenetics and cancer development, 92 (pre)malignant and benign samples, originating from two different collections of cervical samples and related cell lines, were used in this study. These samples include primary carcinomas (n = 22), low- and high-grade cervical intraepithelial neoplasia (CIN1 and CIN2/3 - n = 2/n = 30) and normal tissue (n = 20), as well as control samples (n = 17). Viruses that were detected include phages, adenoviruses, herpesviridae and HPV. HPV, which causes virtually all cervical cancers, was identified in 95% of the carcinomas, 100% of the CIN2/3 samples, both CIN1 samples and in 55% of the normal samples. Comparing the amount of mapped fragments on HPV for each HPV-infected sample yielded a significant difference between normal samples and carcinomas or CIN2/3 samples (adjusted p-values resp. <10(-5), <10(-5)), reflecting different viral loads and/or methylation degrees in non-normal samples. Fragments originating from different HPV types could be distinguished and were independently validated by PCR-based assays in 71% of the detections. In conclusion, although limited by the a priori knowledge of viral reference genome sequences, the proposed methodology can provide a first confined but substantial insight into the presence, concentration and types of methylated viral sequences in MBD-seq data at low additional cost.
Keywords: DNA-methylation; MBD-seq; bioinformatics; cervical cancer; epigenomics; human papillomavirus; next generation sequencing; viruses.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

