Systems biologic analysis of T regulatory cells genetic pathways in murine primary biliary cirrhosis
- PMID: 25701076
- PMCID: PMC4829434
- DOI: 10.1016/j.jaut.2015.01.011
Systems biologic analysis of T regulatory cells genetic pathways in murine primary biliary cirrhosis
Abstract
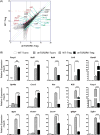
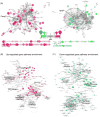
CD4(+)Foxp3(+) regulatory T cells (Tregs) play a non-redundant role in control of excessive immune responses, and defects in Tregs have been shown both in patients and murine models of primary biliary cirrhosis (PBC), a progressive autoimmune biliary disease. Herein, we took advantage of a murine model of PBC, the dominant negative transforming growth factor β receptor II (dnTGFβRII) mice, to assess Treg genetic defects and their functional effects in PBC. By using high-resolution microarrays with verification by PCR and protein expression, we found profound and wide-ranging differences between dnTGFβRII and normal, wild type Tregs. Critical transcription factors were down-regulated including Eos, Ahr, Klf2, Foxp1 in dnTGFβRII Tregs. Functionally, dnTGFβRII Tregs expressed an activated, pro-inflammatory phenotype with upregulation of Ccl5, Granzyme B and IFN-γ. Genetic pathway analysis suggested that the primary effect of loss of TGFβ pathway signaling was to down regulate immune regulatory processes, with a secondary upregulation of inflammatory processes. These findings provide new insights into T regulatory genetic defects; aberrations of the identified genes or genetic pathways should be investigated in human PBC Tregs. This approach which takes advantage of biologic pathway analysis illustrates the ability to identify genes/pathways that are affected both independently and dependent on abnormalities in TGFβ signaling. Such approaches will become increasingly useful in human autoimmunity.
Keywords: Cholangitis; Primary biliary cirrhosis; Regulatory T cells; Transcription profile and pathway analysis.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
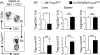




Similar articles
-
Murine autoimmune cholangitis requires two hits: cytotoxic KLRG1(+) CD8 effector cells and defective T regulatory cells.J Autoimmun. 2014 May;50:123-34. doi: 10.1016/j.jaut.2014.01.034. Epub 2014 Feb 18. J Autoimmun. 2014. PMID: 24556277 Free PMC article.
-
Natural killer T cells exacerbate liver injury in a transforming growth factor beta receptor II dominant-negative mouse model of primary biliary cirrhosis.Hepatology. 2008 Feb;47(2):571-80. doi: 10.1002/hep.22052. Hepatology. 2008. PMID: 18098320
-
Liver-targeted and peripheral blood alterations of regulatory T cells in primary biliary cirrhosis.Hepatology. 2006 Apr;43(4):729-37. doi: 10.1002/hep.21123. Hepatology. 2006. PMID: 16557534
-
Animal models of primary biliary cirrhosis.Semin Liver Dis. 2014 Aug;34(3):285-96. doi: 10.1055/s-0034-1383728. Epub 2014 Jul 24. Semin Liver Dis. 2014. PMID: 25057952 Review.
-
Regulatory T cells: Mechanisms of suppression and impairment in autoimmune liver disease.IUBMB Life. 2015 Feb;67(2):88-97. doi: 10.1002/iub.1349. Epub 2015 Apr 8. IUBMB Life. 2015. PMID: 25850692 Review.
Cited by
-
Proposed therapies in primary biliary cholangitis.Expert Rev Gastroenterol Hepatol. 2016;10(3):371-382. doi: 10.1586/17474124.2016.1121810. Epub 2016 Jan 6. Expert Rev Gastroenterol Hepatol. 2016. PMID: 26577047 Free PMC article. Review.
-
Adaptive immunity in the liver.Cell Mol Immunol. 2016 May;13(3):354-68. doi: 10.1038/cmi.2016.4. Epub 2016 Mar 21. Cell Mol Immunol. 2016. PMID: 26996069 Free PMC article. Review.
-
Regulatory T cells in autoimmune hepatitis: an updated overview.J Autoimmun. 2021 May;119:102619. doi: 10.1016/j.jaut.2021.102619. Epub 2021 Feb 27. J Autoimmun. 2021. PMID: 33652348 Free PMC article. Review.
-
Block of both TGF-β and IL-2 signaling impedes Neurophilin-1+ regulatory T cell and follicular regulatory T cell development.Cell Death Dis. 2016 Oct 27;7(10):e2439. doi: 10.1038/cddis.2016.348. Cell Death Dis. 2016. PMID: 27787514 Free PMC article.
-
Liver-resident NK cells suppress autoimmune cholangitis and limit the proliferation of CD4+ T cells.Cell Mol Immunol. 2020 Feb;17(2):178-189. doi: 10.1038/s41423-019-0199-z. Epub 2019 Mar 15. Cell Mol Immunol. 2020. PMID: 30874628 Free PMC article.
References
-
- Gershwin ME, Mackay IR, Sturgess A, Coppel RL. Identification and specificity of a cDNA encoding the 70 kd mitochondrial antigen recognized in primary biliary cirrhosis. Journal of immunology. 1987;138:3525–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

