Dystrophin genotype-cardiac phenotype correlations in Duchenne and Becker muscular dystrophies using cardiac magnetic resonance imaging
- PMID: 25702278
- PMCID: PMC5568575
- DOI: 10.1016/j.amjcard.2015.01.030
Dystrophin genotype-cardiac phenotype correlations in Duchenne and Becker muscular dystrophies using cardiac magnetic resonance imaging
Abstract
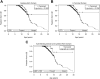
Duchenne and Becker muscular dystrophies are caused by mutations in dystrophin. Cardiac manifestations vary broadly, making prognosis difficult. Current dystrophin genotype-cardiac phenotype correlations are limited. For skeletal muscle, the reading-frame rule suggests in-frame mutations tend to yield milder phenotypes. We performed dystrophin genotype-cardiac phenotype correlations using a protein-effect model and cardiac magnetic resonance imaging. A translational model was applied to patient-specific deletion, indel, and nonsense mutations to predict exons and protein domains present within truncated dystrophin protein. Patients were dichotomized into predicted present and predicted absent groups for exons and protein domains of interest. Development of myocardial fibrosis (represented by late gadolinium enhancement [LGE]) and depressed left ventricular ejection fraction (LVEF) were compared. Patients (n = 274) with predicted present cysteine-rich domain (CRD), C-terminal domain (CTD), and both the N-terminal actin-binding and cysteine-rich domains (ABD1 + CRD) had a decreased risk of LGE and trended toward greater freedom from LGE. Patients with predicted present CTD (exactly the same as those with in-frame mutations) and ABD1 + CRD trended toward decreased risk of and greater freedom from depressed LVEF. In conclusion, genotypes previously implicated in altering the dystrophinopathic cardiac phenotype were not significantly related to LGE and depressed LVEF. Patients with predicted present CRD, CTD/in-frame mutations, and ABD1 + CRD trended toward milder cardiac phenotypes, suggesting that the reading-frame rule may be applicable to the cardiac phenotype. Genotype-phenotype correlations may help predict the cardiac phenotype for dystrophinopathic patients and guide future therapies.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures


References
-
- Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. - PubMed
-
- Finsterer J, Stollberger C. The heart in human dystrophinopathies. Cardiology. 2003;99:1–19. - PubMed
-
- Connuck DM, Sleeper LA, Colan SD, Cox GF, Towbin JA, Lowe AM, Wilkinson JD, Orav EJ, Cuniberti L, Salbert BA, Lipshultz SE. Pediatric Cardiomyopathy Registry Study Group. Characteristics and outcomes of cardiomyopathy in children with Duchenne or Becker muscular dystrophy: a comparative study from the Pediatric Cardiomyopathy Registry. Am Heart J. 2008;155:998–1005. - PMC - PubMed
-
- Jefferies JL, Eidem BW, Belmont JW, Craigen WJ, Ware SM, Fernbach SD, Neish SR, Smith EO, Towbin JA. Genetic predictors and remodeling of dilated cardiomyopathy in muscular dystrophy. Circulation. 2005;112:2799–2804. - PubMed
-
- Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

