Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex
- PMID: 25702574
- PMCID: PMC4366288
- DOI: 10.1016/j.cub.2015.01.041
Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex
Abstract
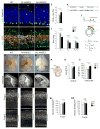
The human neocortex differs from that of other great apes in several notable regards, including altered cell cycle, prolonged corticogenesis, and increased size [1-5]. Although these evolutionary changes most likely contributed to the origin of distinctively human cognitive faculties, their genetic basis remains almost entirely unknown. Highly conserved non-coding regions showing rapid sequence changes along the human lineage are candidate loci for the development and evolution of uniquely human traits. Several studies have identified human-accelerated enhancers [6-14], but none have linked an expression difference to a specific organismal trait. Here we report the discovery of a human-accelerated regulatory enhancer (HARE5) of FZD8, a receptor of the Wnt pathway implicated in brain development and size [15, 16]. Using transgenic mice, we demonstrate dramatic differences in human and chimpanzee HARE5 activity, with human HARE5 driving early and robust expression at the onset of corticogenesis. Similar to HARE5 activity, FZD8 is expressed in neural progenitors of the developing neocortex [17-19]. Chromosome conformation capture assays reveal that HARE5 physically and specifically contacts the core Fzd8 promoter in the mouse embryonic neocortex. To assess the phenotypic consequences of HARE5 activity, we generated transgenic mice in which Fzd8 expression is under control of orthologous enhancers (Pt-HARE5::Fzd8 and Hs-HARE5::Fzd8). In comparison to Pt-HARE5::Fzd8, Hs-HARE5::Fzd8 mice showed marked acceleration of neural progenitor cell cycle and increased brain size. Changes in HARE5 function unique to humans thus alter the cell-cycle dynamics of a critical population of stem cells during corticogenesis and may underlie some distinctive anatomical features of the human brain.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures



Comment in
-
Human evolution: enhancing the brain.Curr Biol. 2015 May 18;25(10):R421-3. doi: 10.1016/j.cub.2015.03.031. Curr Biol. 2015. PMID: 25989083
References
-
- Enard W, Gehre S, Hammerschmidt K, HOlter SM, Blass T, Somel M, BrUckner MK, Schreiweis C, Winter C, Sohr R, et al. A Humanized Version of Foxp2 Affects Cortico-Basal Ganglia Circuits in Mice. Cell. 2009;137:961–971. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

