Evidence that mutation accumulation does not cause aging in Saccharomyces cerevisiae
- PMID: 25702753
- PMCID: PMC4406665
- DOI: 10.1111/acel.12290
Evidence that mutation accumulation does not cause aging in Saccharomyces cerevisiae
Abstract
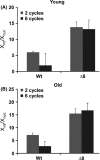
The concept that mutations cause aging phenotypes could not be directly tested previously due to inability to identify age-related mutations in somatic cells and determine their impact on organismal aging. Here, we subjected Saccharomyces cerevisiae to multiple rounds of replicative aging and assessed de novo mutations in daughters of mothers of different age. Mutations did increase with age, but their low numbers, < 1 per lifespan, excluded their causal role in aging. Structural genome changes also had no role. A mutant lacking thiol peroxidases had the mutation rate well above that of wild-type cells, but this did not correspond to the aging pattern, as old wild-type cells with few or no mutations were dying, whereas young mutant cells with many more mutations continued dividing. In addition, wild-type cells lost mitochondrial DNA during aging, whereas shorter-lived mutant cells preserved it, excluding a causal role of mitochondrial mutations in aging. Thus, DNA mutations do not cause aging in yeast. These findings may apply to other damage types, suggesting a causal role of cumulative damage, as opposed to individual damage types, in organismal aging.
Keywords: DNA damage; aging; lifespan; mitochondria; mutations; thiol peroxidase; yeast.
© 2014 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

