Characterization of disulfide bonds by planned digestion and tandem mass spectrometry
- PMID: 25703060
- PMCID: PMC4410109
- DOI: 10.1039/c4mb00688g
Characterization of disulfide bonds by planned digestion and tandem mass spectrometry
Abstract
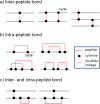
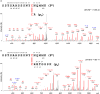
The identification of disulfide bonds provides critical information regarding the structure and function of a protein and is a key aspect in understanding signaling cascades in biological systems. Recent proteomic approaches using digestion enzymes have facilitated the characterization of disulfide-bonds and/or oxidized products from cysteine residues, although these methods have limitations in the application of MS/MS. For example, protein digestion to obtain the native form of disulfide bonds results in short lengths of amino acids, which can cause ambiguous MS/MS analysis due to false positive identifications. In this study we propose a new approach, termed planned digestion, to obtain sufficient amino acid lengths after cleavage for proteomic approaches. Application of the DBond software to planned digestion of specific proteins accurately identified disulfide-linked peptides. RNase A was used as a model protein in this study because the disulfide bonds of this protein have been well characterized. Application of this approach to peptides digested with Asp-N/C (chemical digestion) and trypsin under acid hydrolysis conditions identified the four native disulfide bonds of RNase A. Missed cleavages introduced by trypsin treatment for only 3 hours generated sufficient lengths of amino acids for identification of the disulfide bonds. Analysis using MS/MS successfully showed additional fragmentation patterns that are cleavage products of S-S and C-S bonds of disulfide-linkage peptides. These fragmentation patterns generate thioaldehydes, persulfide, and dehydroalanine. This approach of planned digestion with missed cleavages using the DBond algorithm could be applied to other proteins to determine their disulfide linkage and the oxidation patterns of cysteine residues.
Figures


Similar articles
-
New algorithm for the identification of intact disulfide linkages based on fragmentation characteristics in tandem mass spectra.J Proteome Res. 2010 Jan;9(1):626-35. doi: 10.1021/pr900771r. J Proteome Res. 2010. PMID: 19902913
-
In-Depth Characterization of Protein Disulfide Bonds by Online Liquid Chromatography-Electrochemistry-Mass Spectrometry.J Am Soc Mass Spectrom. 2016 Jan;27(1):50-8. doi: 10.1007/s13361-015-1258-z. Epub 2015 Sep 14. J Am Soc Mass Spectrom. 2016. PMID: 26369777 Free PMC article.
-
Generic Workflow for Mapping of Complex Disulfide Bonds Using In-Source Reduction and Extracted Ion Chromatograms from Data-Dependent Mass Spectrometry.Anal Chem. 2018 Jul 3;90(13):8202-8210. doi: 10.1021/acs.analchem.8b01603. Epub 2018 Jun 22. Anal Chem. 2018. PMID: 29878755
-
Protein disulfide bond determination by mass spectrometry.Mass Spectrom Rev. 2002 May-Jun;21(3):183-216. doi: 10.1002/mas.10025. Mass Spectrom Rev. 2002. PMID: 12476442 Review.
-
Software eyes for protein post-translational modifications.Mass Spectrom Rev. 2015 Mar-Apr;34(2):133-47. doi: 10.1002/mas.21425. Epub 2014 Jun 2. Mass Spectrom Rev. 2015. PMID: 24889695 Review.
Cited by
-
Computational methods in mass spectrometry-based structural proteomics for studying protein structure, dynamics, and interactions.Comput Struct Biotechnol J. 2020 Jun 10;18:1391-1402. doi: 10.1016/j.csbj.2020.06.002. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32637038 Free PMC article. Review.
-
Comprehensive identification of protein disulfide bonds with pepsin/trypsin digestion, Orbitrap HCD and Spectrum Identification Machine.J Proteomics. 2019 Apr 30;198:78-86. doi: 10.1016/j.jprot.2018.12.010. Epub 2018 Dec 14. J Proteomics. 2019. PMID: 30557666 Free PMC article.
-
Wheat-Based Glues in Conservation and Cultural Heritage: (Dis)solving the Proteome of Flour and Starch Pastes and Their Adhering Properties.J Proteome Res. 2024 May 3;23(5):1649-1665. doi: 10.1021/acs.jproteome.3c00804. Epub 2024 Apr 4. J Proteome Res. 2024. PMID: 38574199 Free PMC article.
-
Identification of Sulfenylated Cysteines in Arabidopsis thaliana Proteins Using a Disulfide-Linked Peptide Reporter.Front Plant Sci. 2020 Jul 2;11:777. doi: 10.3389/fpls.2020.00777. eCollection 2020. Front Plant Sci. 2020. PMID: 32714340 Free PMC article.
-
Characterization of Disulfide Linkages in Proteins by 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry.Anal Chem. 2018 Jul 17;90(14):8523-8530. doi: 10.1021/acs.analchem.8b01556. Epub 2018 Jun 28. Anal Chem. 2018. PMID: 29902373 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

