Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways
- PMID: 25703095
- PMCID: PMC4344849
- DOI: 10.1016/j.cell.2015.02.003
Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways
Abstract
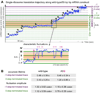
Programmed ribosomal frameshifting produces alternative proteins from a single transcript. -1 frameshifting occurs on Escherichia coli's dnaX mRNA containing a slippery sequence AAAAAAG and peripheral mRNA structural barriers. Here, we reveal hidden aspects of the frameshifting process, including its exact location on the mRNA and its timing within the translation cycle. Mass spectrometry of translated products shows that ribosomes enter the -1 frame from not one specific codon but various codons along the slippery sequence and slip by not just -1 but also -4 or +2 nucleotides. Single-ribosome translation trajectories detect distinctive codon-scale fluctuations in ribosome-mRNA displacement across the slippery sequence, representing multiple ribosomal translocation attempts during frameshifting. Flanking mRNA structural barriers mechanically stimulate the ribosome to undergo back-and-forth translocation excursions, broadly exploring reading frames. Both experiments reveal aborted translation around mutant slippery sequences, indicating that subsequent fidelity checks on newly adopted codon position base pairings lead to either resumed translation or early termination.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures





References
-
- Bretscher MS. Translocation in protein synthesis: a hybrid structure model. Nature. 1968;218:675–677. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

