Nothing to sneeze at: a dynamic and integrative computational model of an influenza A virion
- PMID: 25703376
- PMCID: PMC4353694
- DOI: 10.1016/j.str.2014.12.019
Nothing to sneeze at: a dynamic and integrative computational model of an influenza A virion
Abstract
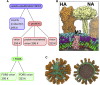
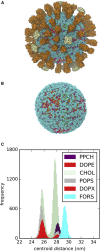
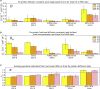
The influenza virus is surrounded by an envelope composed of a lipid bilayer and integral membrane proteins. Understanding the structural dynamics of the membrane envelope provides biophysical insights into aspects of viral function, such as the wide-ranging survival times of the virion in different environments. We have combined experimental data from X-ray crystallography, nuclear magnetic resonance spectroscopy, cryo-electron microscopy, and lipidomics to build a model of the intact influenza A virion. This is the basis of microsecond-scale coarse-grained molecular dynamics simulations of the virion, providing simulations at different temperatures and with varying lipid compositions. The presence of the Forssman glycolipid alters a number of biophysical properties of the virion, resulting in reduced mobility of bilayer lipid and protein species. Reduced mobility in the virion membrane may confer physical robustness to changes in environmental conditions. Our simulations indicate that viral spike proteins do not aggregate and thus are competent for multivalent immunoglobulin G interactions.
Copyright © 2015 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures








Comment in
-
Simulations move toward a cure for viral diseases.Structure. 2015 Mar 3;23(3):439-440. doi: 10.1016/j.str.2015.02.002. Structure. 2015. PMID: 25738384
References
-
- Almeida P.F.F., Vaz W.L.C. Lateral diffusion in membranes. In: Lipowsky R., Sackmann E., editors. Structure and Dynamics of Membranes: From Cells to Vesicles. Vol. 1. North-Holland; 1995. pp. 305–357. (Handbook of Biological Physics).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

