Diversity in gut bacterial community of school-age children in Asia
- PMID: 25703686
- PMCID: PMC4336934
- DOI: 10.1038/srep08397
Diversity in gut bacterial community of school-age children in Asia
Erratum in
-
Author Correction: Diversity in gut bacterial community of school-age children in Asia.Sci Rep. 2019 Apr 25;9(1):6530. doi: 10.1038/s41598-019-42780-z. Sci Rep. 2019. PMID: 31024062 Free PMC article.
Abstract
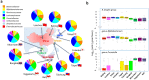
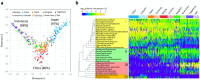
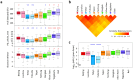
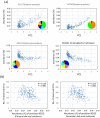
Asia differs substantially among and within its regions populated by diverse ethnic groups, which maintain their own respective cultures and dietary habits. To address the diversity in their gut microbiota, we characterized the bacterial community in fecal samples obtained from 303 school-age children living in urban or rural regions in five countries spanning temperate and tropical areas of Asia. The microbiota profiled for the 303 subjects were classified into two enterotype-like clusters, each driven by Prevotella (P-type) or Bifidobacterium/Bacteroides (BB-type), respectively. Majority in China, Japan and Taiwan harbored BB-type, whereas those from Indonesia and Khon Kaen in Thailand mainly harbored P-type. The P-type microbiota was characterized by a more conserved bacterial community sharing a greater number of type-specific phylotypes. Predictive metagenomics suggests higher and lower activity of carbohydrate digestion and bile acid biosynthesis, respectively, in P-type subjects, reflecting their high intake of diets rich in resistant starch. Random-forest analysis classified their fecal species community as mirroring location of resident country, suggesting eco-geographical factors shaping gut microbiota. In particular, children living in Japan harbored a less diversified microbiota with high abundance of Bifidobacterium and less number of potentially pathogenic bacteria, which may reflect their living environment and unique diet.
Figures



References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

