A generic assay for whole-genome amplification and deep sequencing of enterovirus A71
- PMID: 25704598
- PMCID: PMC4374682
- DOI: 10.1016/j.jviromet.2015.02.011
A generic assay for whole-genome amplification and deep sequencing of enterovirus A71
Abstract
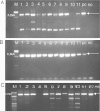
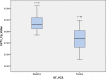
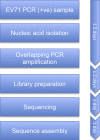
Enterovirus A71 (EV-A71) has emerged as the most important cause of large outbreaks of severe and sometimes fatal hand, foot and mouth disease (HFMD) across the Asia-Pacific region. EV-A71 outbreaks have been associated with (sub)genogroup switches, sometimes accompanied by recombination events. Understanding EV-A71 population dynamics is therefore essential for understanding this emerging infection, and may provide pivotal information for vaccine development. Despite the public health burden of EV-A71, relatively few EV-A71 complete-genome sequences are available for analysis and from limited geographical localities. The availability of an efficient procedure for whole-genome sequencing would stimulate effort to generate more viral sequence data. Herein, we report for the first time the development of a next-generation sequencing based protocol for whole-genome sequencing of EV-A71 directly from clinical specimens. We were able to sequence viruses of subgenogroup C4 and B5, while RNA from culture materials of diverse EV-A71 subgenogroups belonging to both genogroup B and C was successfully amplified. The nature of intra-host genetic diversity was explored in 22 clinical samples, revealing 107 positions carrying minor variants (ranging from 0 to 15 variants per sample). Our analysis of EV-A71 strains sampled in 2013 showed that they all belonged to subgenogroup B5, representing the first report of this subgenogroup in Vietnam. In conclusion, we have successfully developed a high-throughput next-generation sequencing-based assay for whole-genome sequencing of EV-A71 from clinical samples.
Keywords: Deep sequencing; Enterovirus A71; Hand foot and mouth disease; Phylogeny; Picornavirus.
Copyright © 2015 The Authors. Published by Elsevier B.V. All rights reserved.
Figures

References
-
- Chang S.C., Li W.C., Chen G.W., Tsao K.C., Huang C.G., Huang Y.C., Chiu C.H., Kuo C.Y., Tsai K.N., Shih S.R., Lin T.Y. Genetic characterization of enterovirus 71 isolated from patients with severe disease by comparative analysis of complete genomes. J. Med. Virol. 2012;84(6):931–939. - PubMed
-
- Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52(5):696–704. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

