Analysis of Glycosaminoglycans Using Mass Spectrometry
- PMID: 25705143
- PMCID: PMC4334465
- DOI: 10.2174/157016411798220871
Analysis of Glycosaminoglycans Using Mass Spectrometry
Abstract
The glycosaminoglycans (GAGs) are linear polysaccharides expressed on animal cell surfaces and in extracellular matrices. Their biosynthesis is under complex control and confers a domain structure that is essential to their ability to bind to protein partners. Key to understanding the functions of GAGs are methods to determine accurately and rapidly patterns of sulfation, acetylation and uronic acid epimerization that correlate with protein binding or other biological activities. Mass spectrometry (MS) is particularly suitable for the analysis of GAGs for biomedical purposes. Using modern ionization techniques it is possible to accurately determine molecular weights of GAG oligosaccharides and their distributions within a mixture. Methods for direct interfacing with liquid chromatography have been developed to permit online mass spectrometric analysis of GAGs. New tandem mass spectrometric methods for fine structure determination of GAGs are emerging. This review summarizes MS-based approaches for analysis of GAGs, including tissue extraction and chromatographic methods compatible with LC/MS and tandem MS.
Keywords: Mass spectrometry; chondroitin sulfate; dermatan sulfate; glycosaminoglycan; heparan sulfate; heparin; hyaluronan; keratan sulfate; proteoglycan.
Figures

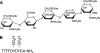
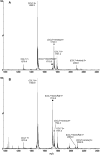
References
-
- Perrimon N, Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature. 2000;404:725–728. - PubMed
-
- Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. - PubMed
-
- Bulow HE, Hobert O. The Molecular Diversity of Glycosaminoglycans Shapes Animal Development. Annu Rev Cell Dev Biol. 2006;22:375–407. - PubMed
-
- Dhoot GK, Gustafsson MK, Ai X, Sun W, Standiford DM, Emerson CP., Jr Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293:1663–1666. - PubMed
-
- Hwang HY, Olson SK, Esko JD, Horvitz HR. Caenorhabditis elegans early embryogenesis and vulval morphogenesis require chondroitin biosynthesis. Nature. 2003;423:439–443. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
