Noncanonical secondary structure stabilizes mitochondrial tRNA(Ser(UCN)) by reducing the entropic cost of tertiary folding
- PMID: 25705930
- PMCID: PMC4399864
- DOI: 10.1021/ja5130308
Noncanonical secondary structure stabilizes mitochondrial tRNA(Ser(UCN)) by reducing the entropic cost of tertiary folding
Abstract
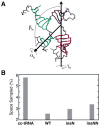
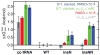
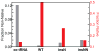
Mammalian mitochondrial tRNA(Ser(UCN)) (mt-tRNA(Ser)) and pyrrolysine tRNA (tRNA(Pyl)) fold to near-canonical three-dimensional structures despite having noncanonical secondary structures with shortened interhelical loops that disrupt the conserved tRNA tertiary interaction network. How these noncanonical tRNAs compensate for their loss of tertiary interactions remains unclear. Furthermore, in human mt-tRNA(Ser), lengthening the variable loop by the 7472insC mutation reduces mt-tRNA(Ser) concentration in vivo through poorly understood mechanisms and is strongly associated with diseases such as deafness and epilepsy. Using simulations of the TOPRNA coarse-grained model, we show that increased topological constraints encoded by the unique secondary structure of wild-type mt-tRNA(Ser) decrease the entropic cost of folding by ∼2.5 kcal/mol compared to canonical tRNA, offsetting its loss of tertiary interactions. Further simulations show that the pathogenic 7472insC mutation disrupts topological constraints and hence destabilizes the mutant mt-tRNA(Ser) by ∼0.6 kcal/mol relative to wild-type. UV melting experiments confirm that insertion mutations lower mt-tRNA(Ser) melting temperature by 6-9 °C and increase the folding free energy by 0.8-1.7 kcal/mol in a largely sequence- and salt-independent manner, in quantitative agreement with our simulation predictions. Our results show that topological constraints provide a quantitative framework for describing key aspects of RNA folding behavior and also provide the first evidence of a pathogenic mutation that is due to disruption of topological constraints.
Figures
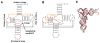
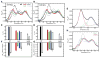
Similar articles
-
The 7472insC mitochondrial DNA mutation impairs the synthesis and extent of aminoacylation of tRNASer(UCN) but not its structure or rate of turnover.J Biol Chem. 2002 Jun 21;277(25):22240-50. doi: 10.1074/jbc.M200338200. Epub 2002 Mar 27. J Biol Chem. 2002. PMID: 11919191
-
A deafness-associated mitochondrial DNA mutation altered the tRNASer(UCN) metabolism and mitochondrial function.Mitochondrion. 2019 May;46:370-379. doi: 10.1016/j.mito.2018.10.001. Epub 2018 Oct 15. Mitochondrion. 2019. PMID: 30336267
-
Topological constraints are major determinants of tRNA tertiary structure and dynamics and provide basis for tertiary folding cooperativity.Nucleic Acids Res. 2014 Oct;42(18):11792-804. doi: 10.1093/nar/gku807. Epub 2014 Sep 12. Nucleic Acids Res. 2014. PMID: 25217593 Free PMC article.
-
[Mutations of mitochondrial tRNASer(UCN) and their connection with hearing loss].Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2017 Feb 10;34(1):128-132. doi: 10.3760/cma.j.issn.1003-9406.2017.01.030. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2017. PMID: 28186612 Review. Chinese.
-
The role of mitochondrial DNA mutations in hearing loss.Biochem Genet. 2013 Aug;51(7-8):588-602. doi: 10.1007/s10528-013-9589-6. Epub 2013 Apr 21. Biochem Genet. 2013. PMID: 23605717 Review.
Cited by
-
Capturing RNA Folding Free Energy with Coarse-Grained Molecular Dynamics Simulations.Sci Rep. 2017 Apr 10;7:45812. doi: 10.1038/srep45812. Sci Rep. 2017. PMID: 28393861 Free PMC article.
-
Naturally Occurring tRNAs With Non-canonical Structures.Front Microbiol. 2020 Oct 21;11:596914. doi: 10.3389/fmicb.2020.596914. eCollection 2020. Front Microbiol. 2020. PMID: 33193279 Free PMC article. Review.
-
The role of structure in regulatory RNA elements.Biosci Rep. 2024 Oct 30;44(10):BSR20240139. doi: 10.1042/BSR20240139. Biosci Rep. 2024. PMID: 39364891 Free PMC article. Review.
-
Intrinsic Properties of tRNA Molecules as Deciphered via Bayesian Network and Distribution Divergence Analysis.Life (Basel). 2018 Feb 8;8(1):5. doi: 10.3390/life8010005. Life (Basel). 2018. PMID: 29419741 Free PMC article.
-
Pressure pushes tRNALys3 into excited conformational states.Proc Natl Acad Sci U S A. 2023 Jun 27;120(26):e2215556120. doi: 10.1073/pnas.2215556120. Epub 2023 Jun 20. Proc Natl Acad Sci U S A. 2023. PMID: 37339210 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

