Identification of microRNA-mRNA dysregulations in paroxysmal atrial fibrillation
- PMID: 25706326
- PMCID: PMC4417399
- DOI: 10.1016/j.ijcard.2015.01.075
Identification of microRNA-mRNA dysregulations in paroxysmal atrial fibrillation
Abstract
Background: The molecular mechanisms underlying the early development of atrial fibrillation (AF) remain poorly understood. Emerging evidence suggests that abnormal epigenetic modulation via microRNAs (miRNAs) might be involved in the pathogenesis of paroxysmal AF (pAF).
Objective: To identify key molecular changes associated with pAF, we conducted state-of-the-art transcriptomic studies to identify the abnormal miRNA-mRNA interactions potentially driving AF development.
Methods: High-quality total RNA including miRNA was isolated from atrial biopsies of age-matched and sex-matched pAF patients and control patients in sinus rhythm (SR; n=4 per group) and used for RNA-sequencing and miRNA microarray. Results were analyzed bioinformatically and validated using quantitative real-time (qRT)-PCR and 3'UTR luciferase reporter assays.
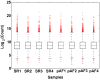
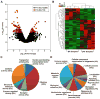
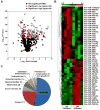
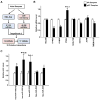
Results: 113 genes and 49 miRNAs were differentially expressed (DE) in pAF versus SR patients. Gene ontology analysis revealed that most of the DE genes were involved in the "gonadotropin releasing hormone receptor pathway" and "p53 pathway". Of these DE genes, bioinformatic analyses identified 23 pairs of putative miRNA-mRNA interactions that were altered in pAF (involving 15 miRNAs and 17 mRNAs). Using qRT-PCR and 3'UTR luciferase reporter assays, the interaction between upregulation of miR-199a-5p and downregulation of FKBP5 was confirmed in samples from pAF patients.
Conclusion: Our combined transcriptomic analysis and miRNA microarray study of atrial samples from pAF patients revealed novel pathways and miRNA-mRNA regulations that may be relevant in the development of pAF. Future studies are required to investigate the potential involvement of the gonadotropin releasing hormone receptor and p53 pathways in AF pathogenesis.
Keywords: Paroxysmal atrial fibrillation; RNA sequencing; Transcriptomic study; microRNA.
Copyright © 2015 Elsevier Ireland Ltd. All rights reserved.
Conflict of interest statement
Figures

References
-
- Jalife J. Mechanisms of persistent atrial fibrillation. Current opinion in cardiology. 2014;29:20–7. - PubMed
-
- Nattel S. Experimental evidence for proarrhythmic mechanisms of antiarrhythmic drugs. Cardiovasc Res. 1998;37:567–77. - PubMed
-
- Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, et al. Reprogramming of the human atrial transcriptome in permanent atrial fibrillation: expression of a ventricular-like genomic signature. Circ Res. 2005;96:1022–9. - PubMed
-
- Kharlap MS, Timofeeva AV, Goryunova LE, Khaspekov GL, Dzemeshkevich SL, Ruskin VV, et al. Atrial appendage transcriptional profile in patients with atrial fibrillation with structural heart diseases. Ann N Y Acad Sci. 2006;1091:205–17. - PubMed
-
- Ohki R, Yamamoto K, Ueno S, Mano H, Misawa Y, Fuse K, et al. Gene expression profiling of human atrial myocardium with atrial fibrillation by DNA microarray analysis. Int J Cardiol. 2005;102:233–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

