Selectivity by small-molecule inhibitors of protein interactions can be driven by protein surface fluctuations
- PMID: 25706586
- PMCID: PMC4338137
- DOI: 10.1371/journal.pcbi.1004081
Selectivity by small-molecule inhibitors of protein interactions can be driven by protein surface fluctuations
Abstract
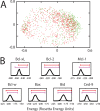
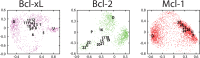
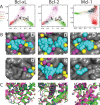
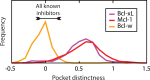
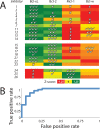
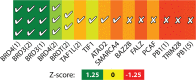
Small-molecules that inhibit interactions between specific pairs of proteins have long represented a promising avenue for therapeutic intervention in a variety of settings. Structural studies have shown that in many cases, the inhibitor-bound protein adopts a conformation that is distinct from its unbound and its protein-bound conformations. This plasticity of the protein surface presents a major challenge in predicting which members of a protein family will be inhibited by a given ligand. Here, we use biased simulations of Bcl-2-family proteins to generate ensembles of low-energy conformations that contain surface pockets suitable for small molecule binding. We find that the resulting conformational ensembles include surface pockets that mimic those observed in inhibitor-bound crystal structures. Next, we find that the ensembles generated using different members of this protein family are overlapping but distinct, and that the activity of a given compound against a particular family member (ligand selectivity) can be predicted from whether the corresponding ensemble samples a complementary surface pocket. Finally, we find that each ensemble includes certain surface pockets that are not shared by any other family member: while no inhibitors have yet been identified to take advantage of these pockets, we expect that chemical scaffolds complementing these "distinct" pockets will prove highly selective for their targets. The opportunity to achieve target selectivity within a protein family by exploiting differences in surface fluctuations represents a new paradigm that may facilitate design of family-selective small-molecule inhibitors of protein-protein interactions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

