Proinflammatory microenvironments within the intestine regulate the differentiation of tissue-resident CD8⁺ T cells responding to infection
- PMID: 25706747
- PMCID: PMC4368475
- DOI: 10.1038/ni.3108
Proinflammatory microenvironments within the intestine regulate the differentiation of tissue-resident CD8⁺ T cells responding to infection
Abstract
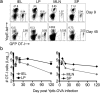
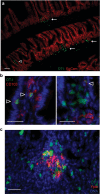
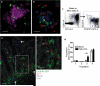
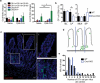
We report that oral infection with Yersinia pseudotuberculosis results in the development of two distinct populations of pathogen-specific CD8(+) tissue-resident memory T cells (TRM cells) in the lamina propria. CD103(-) T cells did not require transforming growth factor-β (TGF-β) signaling but were true resident memory cells. Unlike CD103(+)CD8(+) T cells, which were TGF-β dependent and were scattered in the tissue, CD103(-)CD8(+) T cells clustered with CD4(+) T cells and CX3CR1(+) macrophages and/or dendritic cells around areas of bacterial infection. CXCR3-dependent recruitment of cells to inflamed areas was critical for development of the CD103(-) population and pathogen clearance. Our studies have identified the 'preferential' development of CD103(-) TRM cells in inflammatory microenvironments within the lamina propria and suggest that this subset has a critical role in controlling infection.
Figures




References
-
- Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. - PubMed
-
- Mora JR, et al. Selective imprinting of gut-homing T cells by Peyer's patch dendritic cells. Nature. 2003;424:88–93. - PubMed
-
- Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R. Cutting edge: Gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J. Immunol. 2006;176:2079–2083. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

