Phenotypic responses of differentiated asthmatic human airway epithelial cultures to rhinovirus
- PMID: 25706956
- PMCID: PMC4338293
- DOI: 10.1371/journal.pone.0118286
Phenotypic responses of differentiated asthmatic human airway epithelial cultures to rhinovirus
Abstract
Objectives: Human airway epithelial cells are the principal target of human rhinovirus (HRV), a common cold pathogen that triggers the majority of asthma exacerbations. The objectives of this study were 1) to evaluate an in vitro air liquid interface cultured human airway epithelial cell model for HRV infection, and 2) to identify gene expression patterns associated with asthma intrinsically and/or after HRV infection using this model.
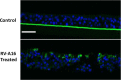
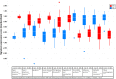
Methods: Air-liquid interface (ALI) human airway epithelial cell cultures were prepared from 6 asthmatic and 6 non-asthmatic donors. The effects of rhinovirus RV-A16 on ALI cultures were compared. Genome-wide gene expression changes in ALI cultures following HRV infection at 24 hours post exposure were further analyzed using RNA-seq technology. Cellular gene expression and cytokine/chemokine secretion were further evaluated by qPCR and a Luminex-based protein assay, respectively.
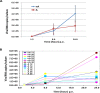
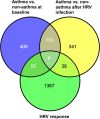
Main results: ALI cultures were readily infected by HRV. RNA-seq analysis of HRV infected ALI cultures identified sets of genes associated with asthma specific viral responses. These genes are related to inflammatory pathways, epithelial structure and remodeling and cilium assembly and function, including those described previously (e.g. CCL5, CXCL10 and CX3CL1, MUC5AC, CDHR3), and novel ones that were identified for the first time in this study (e.g. CCRL1).
Conclusions: ALI-cultured human airway epithelial cells challenged with HRV are a useful translational model for the study of HRV-induced responses in airway epithelial cells, given that gene expression profile using this model largely recapitulates some important patterns of gene responses in patients during clinical HRV infection. Furthermore, our data emphasize that both abnormal airway epithelial structure and inflammatory signaling are two important asthma signatures, which can be further exacerbated by HRV infection.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

