Differential DNA mismatch repair underlies mutation rate variation across the human genome
- PMID: 25707793
- PMCID: PMC4425546
- DOI: 10.1038/nature14173
Differential DNA mismatch repair underlies mutation rate variation across the human genome
Abstract
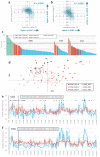
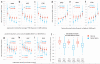
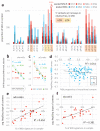
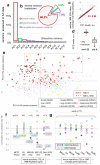
Cancer genome sequencing has revealed considerable variation in somatic mutation rates across the human genome, with mutation rates elevated in heterochromatic late replicating regions and reduced in early replicating euchromatin. Multiple mechanisms have been suggested to underlie this, but the actual cause is unknown. Here we identify variable DNA mismatch repair (MMR) as the basis of this variation. Analysing ∼17 million single-nucleotide variants from the genomes of 652 tumours, we show that regional autosomal mutation rates at megabase resolution are largely stable across cancer types, with differences related to changes in replication timing and gene expression. However, mutations arising after the inactivation of MMR are no longer enriched in late replicating heterochromatin relative to early replicating euchromatin. Thus, differential DNA repair and not differential mutation supply is the primary cause of the large-scale regional mutation rate variation across the human genome.
Figures



References
-
- Hodgkinson A, Chen Y, Eyre-Walker A. The large-scale distribution of somatic mutations in cancer genomes. Hum. Mutat. 2012 doi:10.1002/humu.21616. - PubMed
-
- Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. - PubMed
-
- Woo YH, Li W-H. DNA replication timing and selection shape the landscape of nucleotide variation in cancer genomes. Nat. Commun. 2012;3:1004. - PubMed
Methods’ references
-
- Saunders CT, et al. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics. 2012;28:1811–1817. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

