Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity
- PMID: 25707795
- PMCID: PMC4359072
- DOI: 10.1038/nature14237
Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity
Abstract
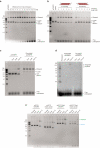
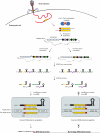
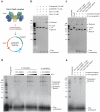
Bacteria and archaea insert spacer sequences acquired from foreign DNAs into CRISPR loci to generate immunological memory. The Escherichia coli Cas1-Cas2 complex mediates spacer acquisition in vivo, but the molecular mechanism of this process is unknown. Here we show that the purified Cas1-Cas2 complex integrates oligonucleotide DNA substrates into acceptor DNA to yield products similar to those generated by retroviral integrases and transposases. Cas1 is the catalytic subunit and Cas2 substantially increases integration activity. Protospacer DNA with free 3'-OH ends and supercoiled target DNA are required, and integration occurs preferentially at the ends of CRISPR repeats and at sequences adjacent to cruciform structures abutting AT-rich regions, similar to the CRISPR leader sequence. Our results demonstrate the Cas1-Cas2 complex to be the minimal machinery that catalyses spacer DNA acquisition and explain the significance of CRISPR repeats in providing sequence and structural specificity for Cas1-Cas2-mediated adaptive immunity.
Figures

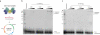







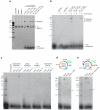

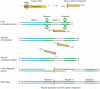
Comment in
-
Microbiology: How bacteria get spacers from invaders.Nature. 2015 Mar 12;519(7542):166-7. doi: 10.1038/nature14204. Epub 2015 Feb 18. Nature. 2015. PMID: 25707799 No abstract available.
References
-
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi:10.1126/science.1138140. - PubMed
-
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of molecular evolution. 2005;60:174–182. doi:10.1007/s00239-004-0046-3. - PubMed
-
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi:10.1099/mic.0.28048-0. - PubMed
-
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi:10.1099/mic.0.27437-0. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

